|
||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for EIF4G1 |
| Basic gene info. | Gene symbol | EIF4G1 |
| Gene name | eukaryotic translation initiation factor 4 gamma, 1 | |
| Synonyms | EIF-4G1|EIF4F|EIF4G|EIF4GI|P220|PARK18 | |
| Cytomap | UCSC genome browser: 3q27.1 | |
| Genomic location | chr3 :184032282-184053146 | |
| Type of gene | protein-coding | |
| RefGenes | NM_001194946.1, NM_001194947.1,NM_001291157.1,NM_004953.4,NM_182917.4, NM_198241.2,NM_198242.2,NM_198244.2, | |
| Ensembl id | ENSG00000114867 | |
| Description | EIF4-gammaeIF-4-gamma 1eucaryotic translation initiation factor 4Geukaryotic translation initiation factor 4 gamma 1 | |
| Modification date | 20141207 | |
| dbXrefs | MIM : 600495 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000114867 | ||
| HPRD : 06774 | ||
| Vega : OTTHUMG00000156784 | ||
| Protein | UniProt: Q04637 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_EIF4G1 | |
| BioGPS: 1981 | ||
| Gene Expression Atlas: ENSG00000114867 | ||
| The Human Protein Atlas: ENSG00000114867 | ||
| Pathway | NCI Pathway Interaction Database: EIF4G1 | |
| KEGG: EIF4G1 | ||
| REACTOME: EIF4G1 | ||
| ConsensusPathDB | ||
| Pathway Commons: EIF4G1 | ||
| Metabolism | MetaCyc: EIF4G1 | |
| HUMANCyc: EIF4G1 | ||
| Regulation | Ensembl's Regulation: ENSG00000114867 | |
| miRBase: chr3 :184,032,282-184,053,146 | ||
| TargetScan: NM_001194946 | ||
| cisRED: ENSG00000114867 | ||
| Context | iHOP: EIF4G1 | |
| cancer metabolism search in PubMed: EIF4G1 | ||
| UCL Cancer Institute: EIF4G1 | ||
| Assigned class in ccmGDB | A - This gene has a literature evidence and it belongs to cancer gene. | |
| References showing role of EIF4G1 in cancer cell metabolism | 1. de la Parra C, Borrero-Garcia LD, Cruz-Collazo A, Schneider RJ, Dharmawardhane S (2015) Equol, an isoflavone metabolite, regulates cancer cell viability and protein synthesis initiation via c-Myc and eIF4G. J Biol Chem 290: 6047-6057. doi: 10.1074/jbc.M114.617415. pmid: 4358247. go to article 2. Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, et al. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109-113. doi: 10.1038/nature11083. pmid: 3347774. go to article 3. Liang S, Zhou Y, Chen Y, Ke G, Wen H, et al. (2014) Decreased expression of EIF4A1 after preoperative brachytherapy predicts better tumor-specific survival in cervical cancer. Int J Gynecol Cancer 24: 908-915. doi: 10.1097/IGC.0000000000000152. go to article 4. Badura M, Braunstein S, Zavadil J, Schneider RJ (2012) DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci U S A 109: 18767-18772. doi: 10.1073/pnas.1203853109. pmid: 3503184. go to article | |
| Top |
| Phenotypic Information for EIF4G1(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: EIF4G1 |
| Familial Cancer Database: EIF4G1 | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
| . | ||||||||||||||
Oncogene 1 | Significant driver gene in | |||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| REACTOME_METABOLISM_OF_PROTEINS REACTOME_METABOLISM_OF_MRNA REACTOME_METABOLISM_OF_RNA | |
| OMIM | 600495; gene. 600495; gene. 614251; phenotype. 614251; phenotype. |
| Orphanet | 2828; Young adult-onset Parkinsonism. 2828; Young adult-onset Parkinsonism. |
| Disease | KEGG Disease: EIF4G1 |
| MedGen: EIF4G1 (Human Medical Genetics with Condition) | |
| ClinVar: EIF4G1 | |
| Phenotype | MGI: EIF4G1 (International Mouse Phenotyping Consortium) |
| PhenomicDB: EIF4G1 | |
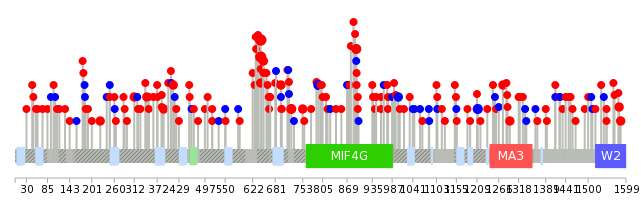
| Mutations for EIF4G1 |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| There's no structural variation information in COSMIC data for this gene. |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows EIF4G1 related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| BF734084 | EIF4G1 | 49 | 166 | 3 | 184045002 | 184045120 | EIF4G1 | 164 | 217 | 3 | 184039685 | 184039738 | |
| AW859459 | EIF4G1 | 6 | 506 | 3 | 184039544 | 184040401 | TPM2 | 501 | 673 | 9 | 35685510 | 35685776 | |
| AI205494 | RABEP1 | 3 | 189 | 17 | 5281795 | 5281981 | EIF4G1 | 188 | 537 | 3 | 184039589 | 184040202 | |
| AI700104 | RABEP1 | 15 | 200 | 17 | 5281795 | 5281981 | EIF4G1 | 199 | 401 | 3 | 184039745 | 184040202 | |
| AW859452 | EIF4G1 | 1 | 503 | 3 | 184039544 | 184040401 | TPM2 | 498 | 663 | 9 | 35685514 | 35685776 | |
| AW859462 | EIF4G1 | 7 | 455 | 3 | 184039604 | 184040401 | TPM2 | 450 | 501 | 9 | 35685725 | 35685776 | |
| BF696853 | RNU6-1 | 1 | 107 | 3 | 180949525 | 180949632 | EIF4G1 | 106 | 625 | 3 | 184038425 | 184039443 | |
| AI744700 | PPP1R14A | 1 | 165 | 19 | 38741882 | 38742046 | EIF4G1 | 161 | 430 | 3 | 184046457 | 184049153 | |
| AU133771 | EIF4G1 | 10 | 361 | 3 | 184040396 | 184040965 | EIF4G1 | 359 | 762 | 3 | 184038480 | 184039375 | |
| BQ212371 | EIF4G1 | 1 | 104 | 3 | 184042831 | 184043088 | EIF4G1 | 98 | 720 | 3 | 184042116 | 184043724 | |
| BQ016901 | UCP2 | 19 | 265 | 11 | 73685717 | 73685963 | EIF4G1 | 257 | 399 | 3 | 184045158 | 184045437 | |
| DA536097 | MYO18A | 1 | 473 | 17 | 27435487 | 27435960 | EIF4G1 | 464 | 520 | 3 | 184043146 | 184043307 | |
| AI656047 | RABEP1 | 3 | 189 | 17 | 5281795 | 5281981 | EIF4G1 | 188 | 635 | 3 | 184039494 | 184040202 | |
| BQ009628 | EIF4G1 | 17 | 213 | 3 | 184052945 | 184053141 | MCC | 212 | 728 | 5 | 112379256 | 112389659 | |
| BE501999 | RABEP1 | 3 | 189 | 17 | 5281795 | 5281981 | EIF4G1 | 188 | 676 | 3 | 184039456 | 184040202 | |
| DA453269 | EIF4G1 | 1 | 140 | 3 | 184035272 | 184037517 | EIF4G1 | 129 | 567 | 3 | 184033920 | 184043666 | |
| F00479 | MIR548N | 2 | 71 | 2 | 179484435 | 179484504 | EIF4G1 | 68 | 268 | 3 | 184046459 | 184049086 | |
| BE466786 | RABEP1 | 3 | 189 | 17 | 5281795 | 5281981 | EIF4G1 | 188 | 633 | 3 | 184039498 | 184040202 | |
| BE077006 | EIF4G1 | 14 | 402 | 3 | 184052077 | 184052470 | HELLS | 394 | 632 | 10 | 96326010 | 96326248 | |
| BE466214 | RABEP1 | 3 | 189 | 17 | 5281795 | 5281981 | EIF4G1 | 188 | 670 | 3 | 184039462 | 184040202 | |
| AK098133 | RAP1A | 1 | 519 | 1 | 112193133 | 112193650 | EIF4G1 | 518 | 2322 | 3 | 184045905 | 184052842 | |
| Top |
| There's no copy number variation information in COSMIC data for this gene. |
| Top |
|
 |
| Top |
| Stat. for Non-Synonymous SNVs (# total SNVs=137) | (# total SNVs=34) |
 | 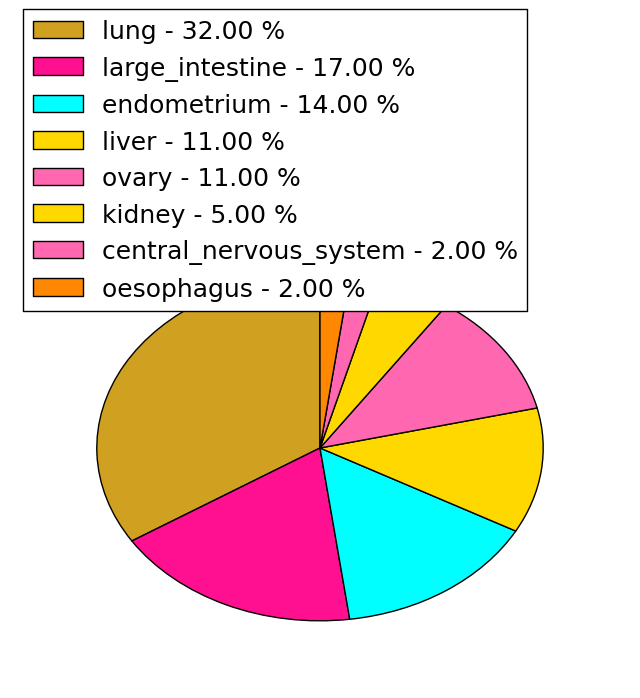 |
(# total SNVs=3) | (# total SNVs=0) |
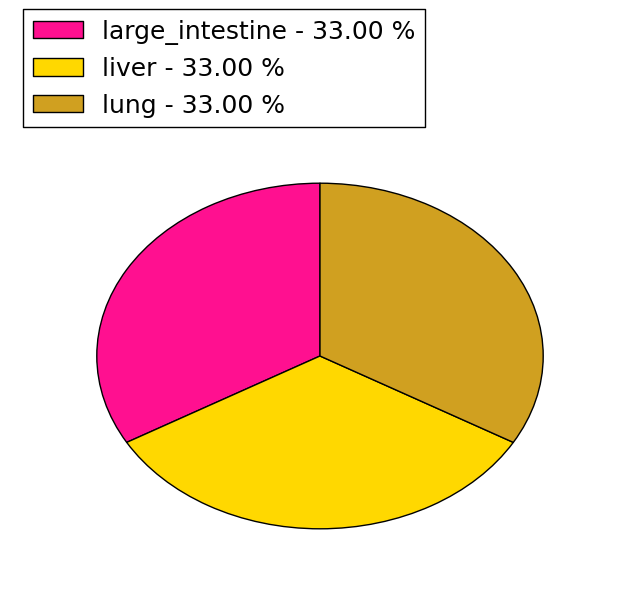 |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr3:184040741-184040741 | p.K643R | 13 |
| chr3:184045208-184045208 | p.T1211T | 3 |
| chr3:184041675-184041675 | p.L794L | 3 |
| chr3:184038427-184038427 | p.R183* | 3 |
| chr3:184041274-184041274 | p.E723K | 3 |
| chr3:184042723-184042723 | p.R893W | 3 |
| chr3:184041028-184041028 | p.P696L | 2 |
| chr3:184041364-184041364 | p.D753N | 2 |
| chr3:184052638-184052638 | p.S1581F | 2 |
| chr3:184040889-184040889 | p.R650W | 2 |
| Top |
|
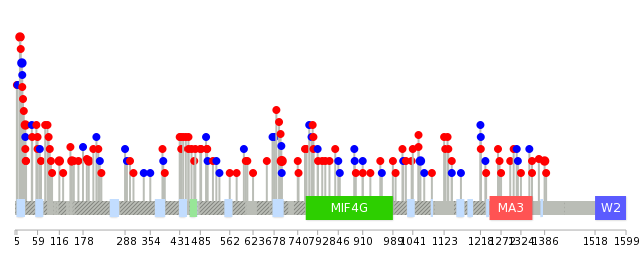 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 5 | 2 | 2 | 16 | 2 | 7 | 1 | 5 | 1 | 17 | 14 | 5 | 1 | 13 | 24 | 1 | 13 | |||
| # mutation | 5 | 2 | 3 | 17 | 2 | 9 | 1 | 5 | 1 | 22 | 17 | 5 | 1 | 15 | 30 | 1 | 18 | |||
| nonsynonymous SNV | 3 | 2 | 3 | 13 | 1 | 7 | 4 | 1 | 12 | 13 | 2 | 1 | 11 | 20 | 1 | 14 | ||||
| synonymous SNV | 2 | 4 | 1 | 2 | 1 | 1 | 10 | 4 | 3 | 4 | 10 | 4 |
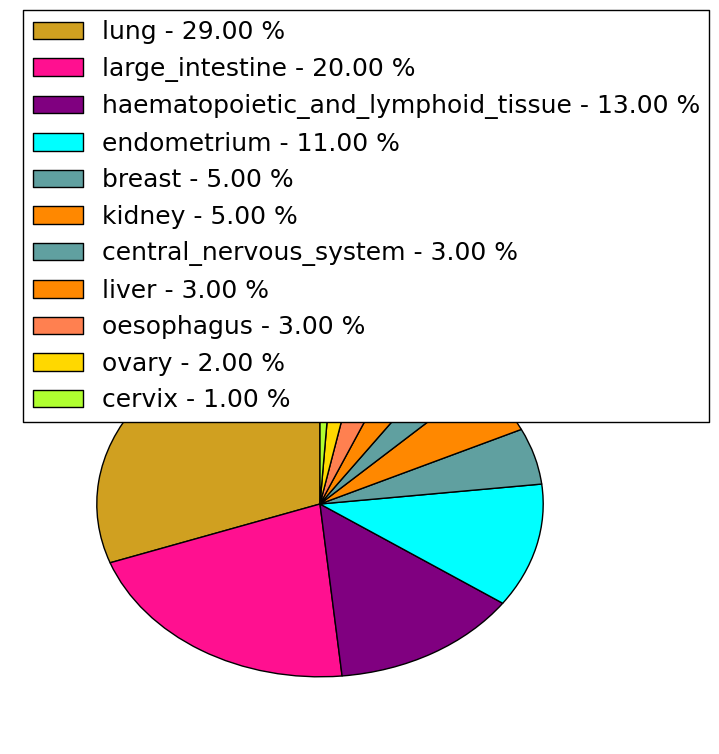
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr3:184042723 | p.R698W,EIF4G1 | 3 |
| chr3:184045480 | p.T27M,EIF4G1 | 2 |
| chr3:184039537 | p.S1386F,EIF4G1 | 2 |
| chr3:184052538 | p.E193K,EIF4G1 | 2 |
| chr3:184038752 | p.L1061L,EIF4G1 | 2 |
| chr3:184039307 | p.R13C,EIF4G1 | 2 |
| chr3:184052638 | p.R116H,EIF4G1 | 2 |
| chr3:184039405 | p.L149F,EIF4G1 | 2 |
| chr3:184037581 | p.A1353T,EIF4G1 | 2 |
| chr3:184044407 | p.S1371G,EIF4G1 | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
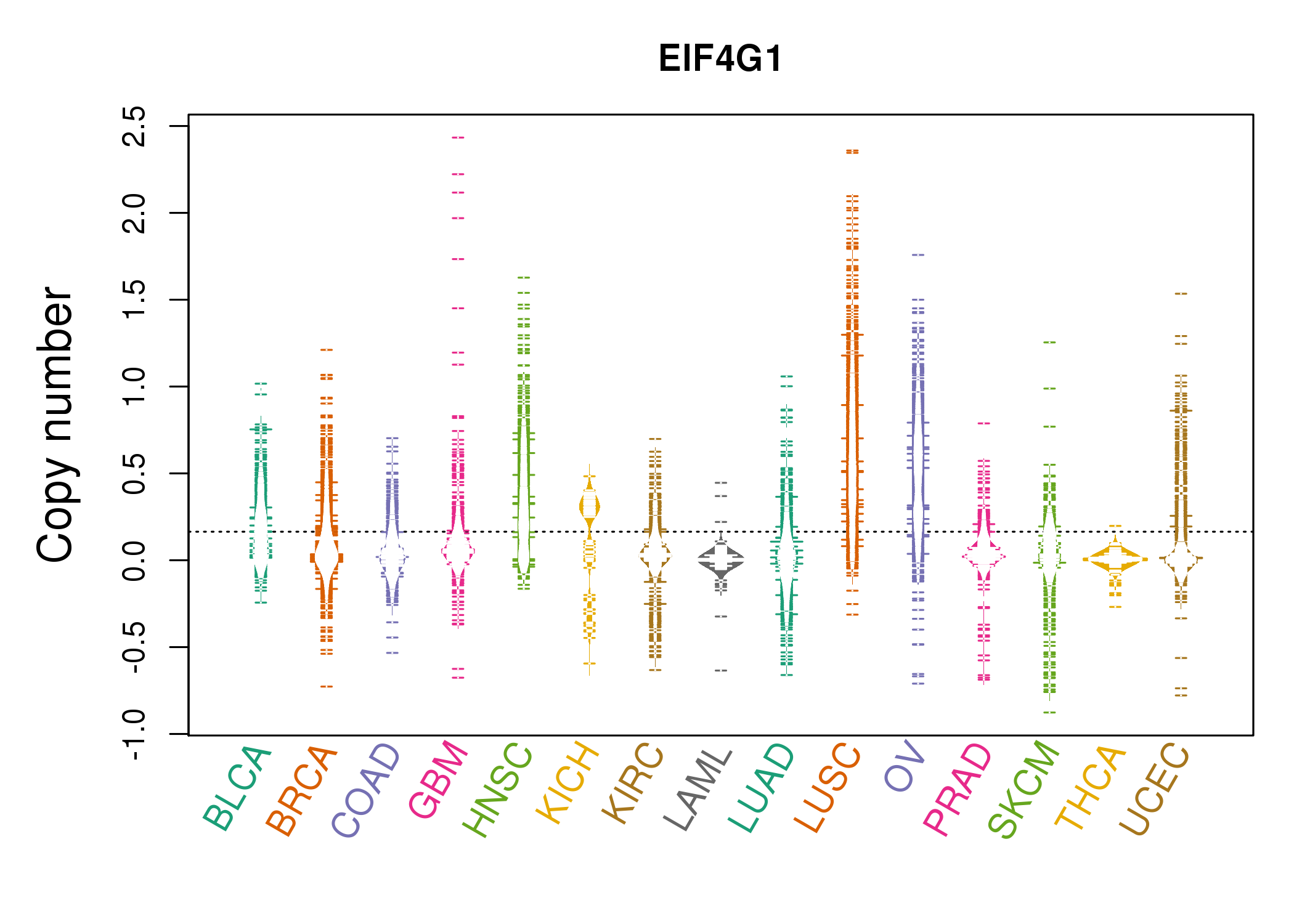 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
| Gene Expression for EIF4G1 |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
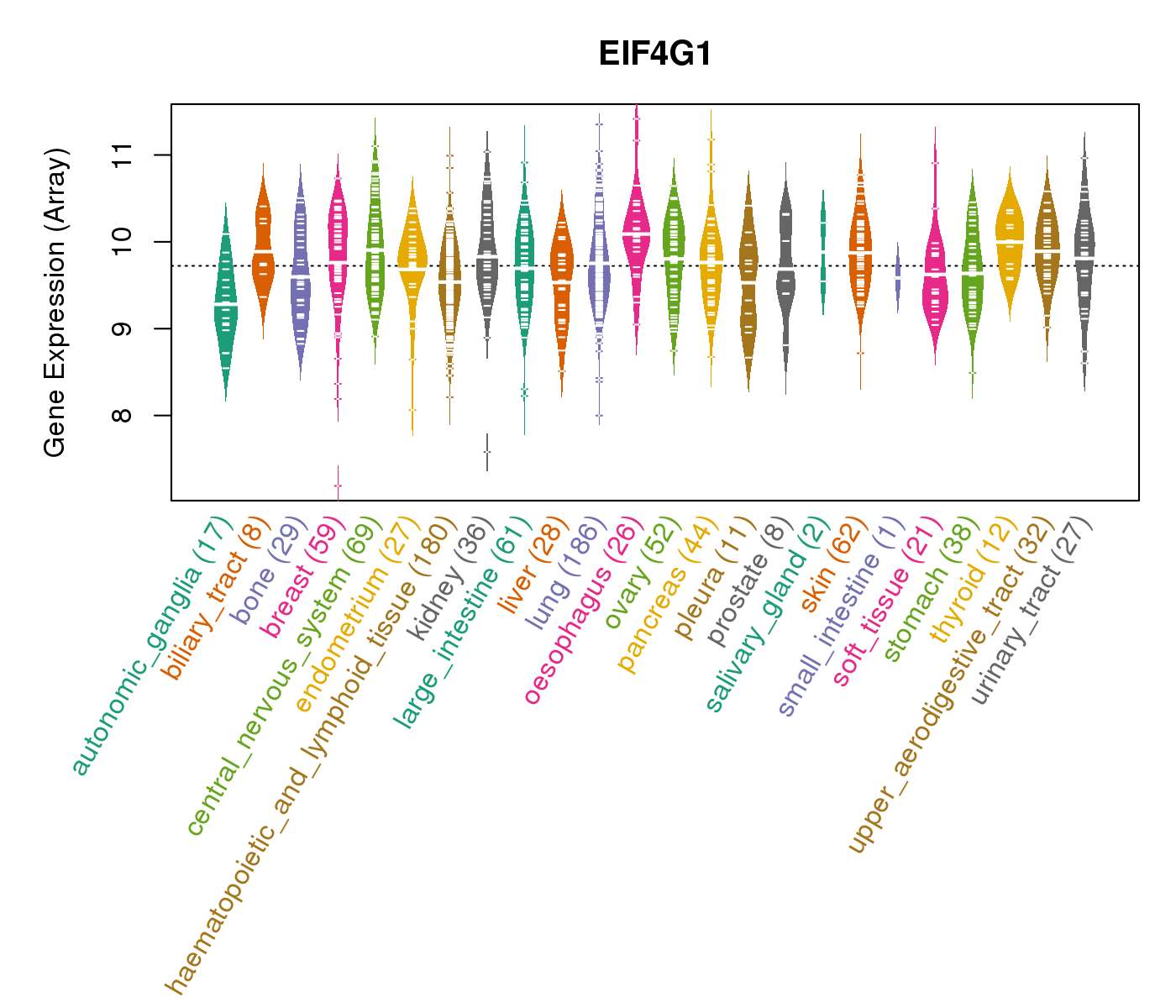 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
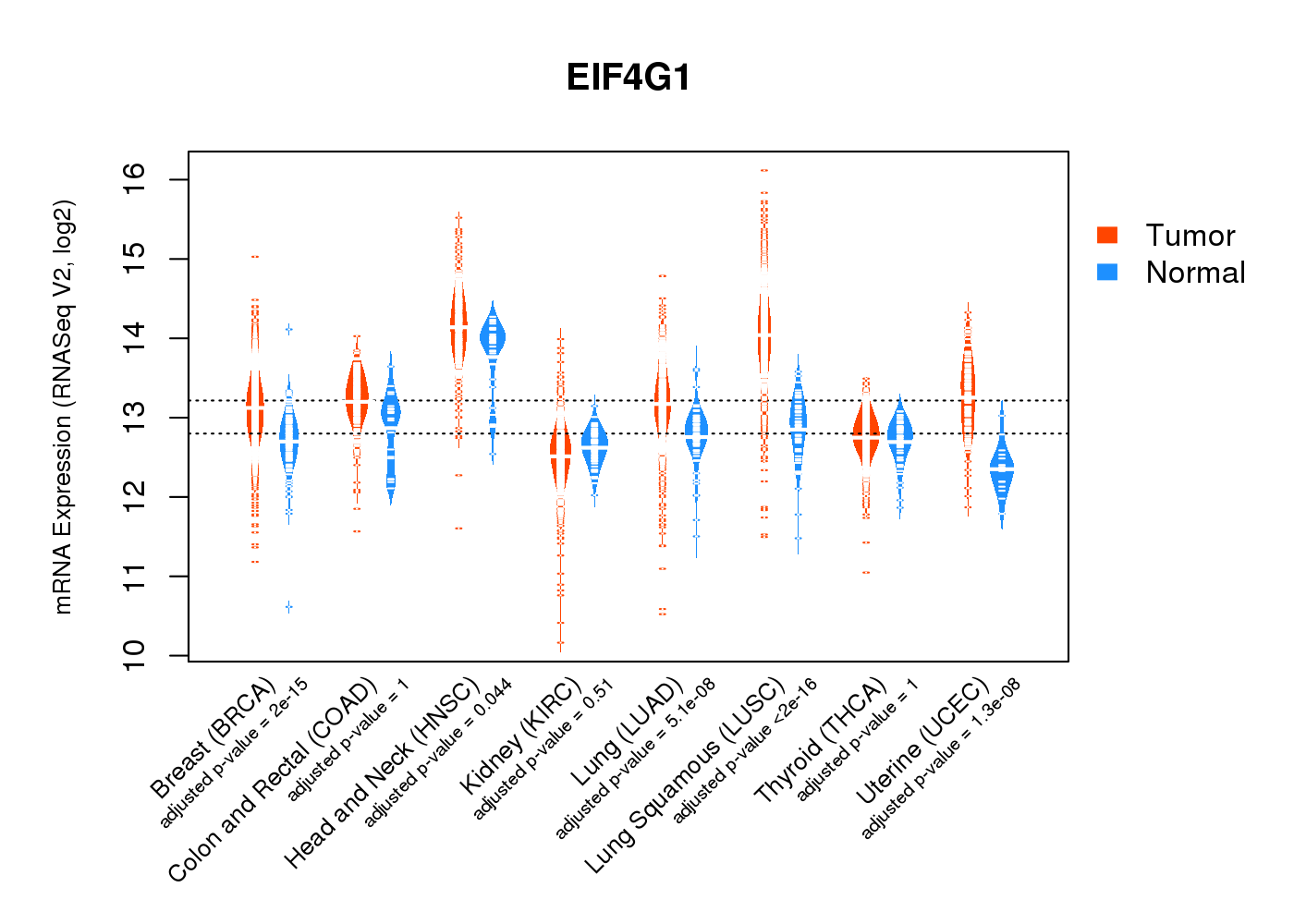 |
| Top |
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
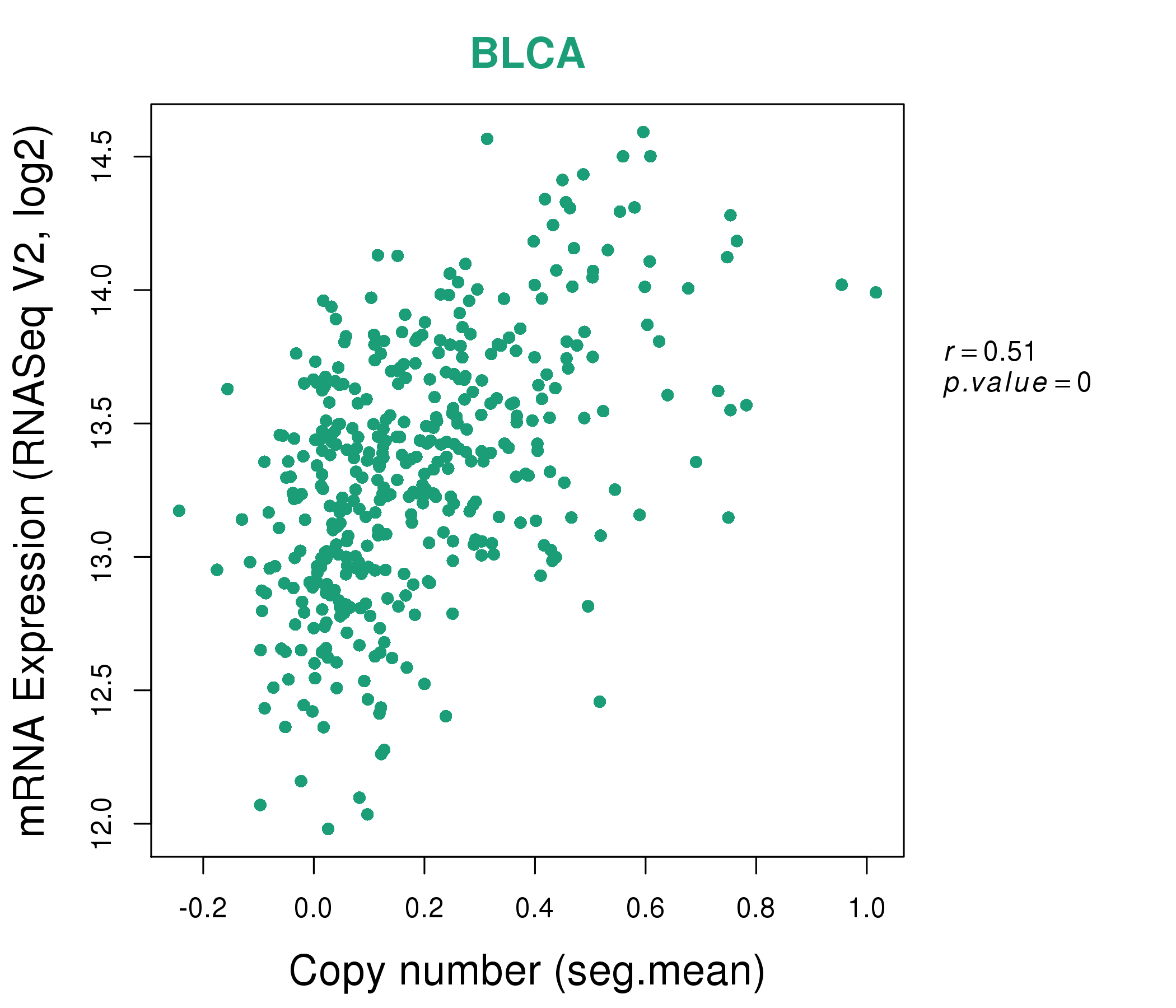 |
|
 |
|
| Top |
| Gene-Gene Network Information |
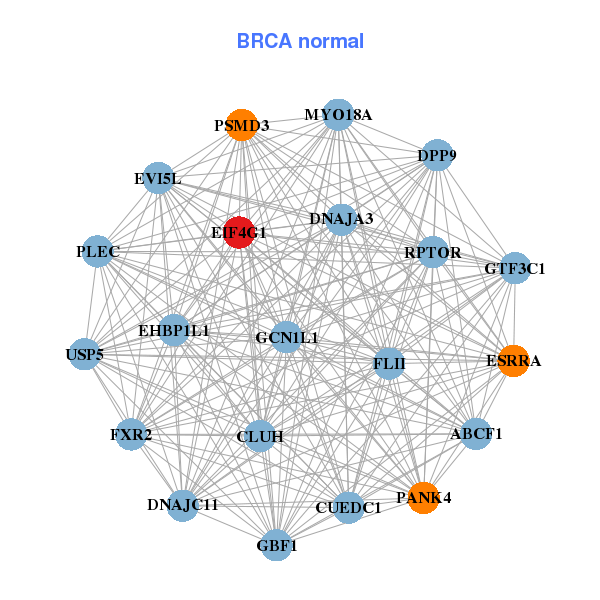
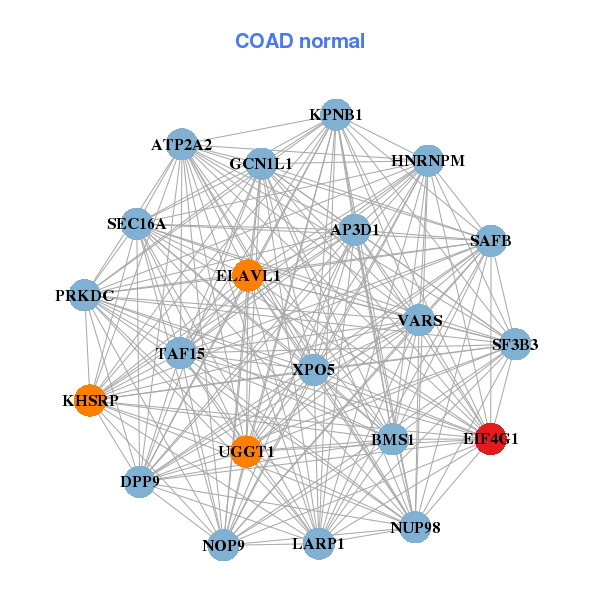
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
 |
| ||||
| ABCF3,AP2M1,DCUN1D1,DVL3,EIF4G1,FBXO45,FXR1, INCENP,KIAA0226,LSG1,MCCC1,OPA1,PCYT1A,POLR1A, PSMD2,RFC4,RNF168,SENP5,TBCCD1,TFRC,YEATS2 | ABCF1,CUEDC1,DNAJA3,DNAJC11,DPP9,EHBP1L1,EIF4G1, ESRRA,EVI5L,FLII,FXR2,GBF1,GCN1L1,GTF3C1, CLUH,MYO18A,PANK4,PLEC,PSMD3,RPTOR,USP5 | ||||
 |
| ||||
| ARID1A,ATP2A2,PRRC2A,NOP9,CHD4,EIF4G1,EP400, GCN1L1,HCFC1,INCENP,KMT2B___KMT2D,NSD1,PIK3R4,POLR3A, PSMD2,RAD54L2,RNF168,SPEN,VPRBP,YEATS2,YLPM1 | AP3D1,ATP2A2,BMS1,NOP9,DPP9,EIF4G1,ELAVL1, GCN1L1,HNRNPM,KHSRP,KPNB1,LARP1,NUP98,PRKDC, SAFB,SEC16A,SF3B3,TAF15,UGGT1,VARS,XPO5 |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for EIF4G1 |
| DB Category | DB Name | DB's ID and Url link |
| Chemistry | BindingDB | Q04637; -. |
| Chemistry | BindingDB | Q04637; -. |
| Organism-specific databases | PharmGKB | PA27722; -. |
| Organism-specific databases | PharmGKB | PA27722; -. |
| Organism-specific databases | CTD | 1981; -. |
| Organism-specific databases | CTD | 1981; -. |
| * Gene Centered Interaction Network. |
 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB00877 | eukaryotic translation initiation factor 4 gamma, 1 | approved; investigational | Sirolimus | 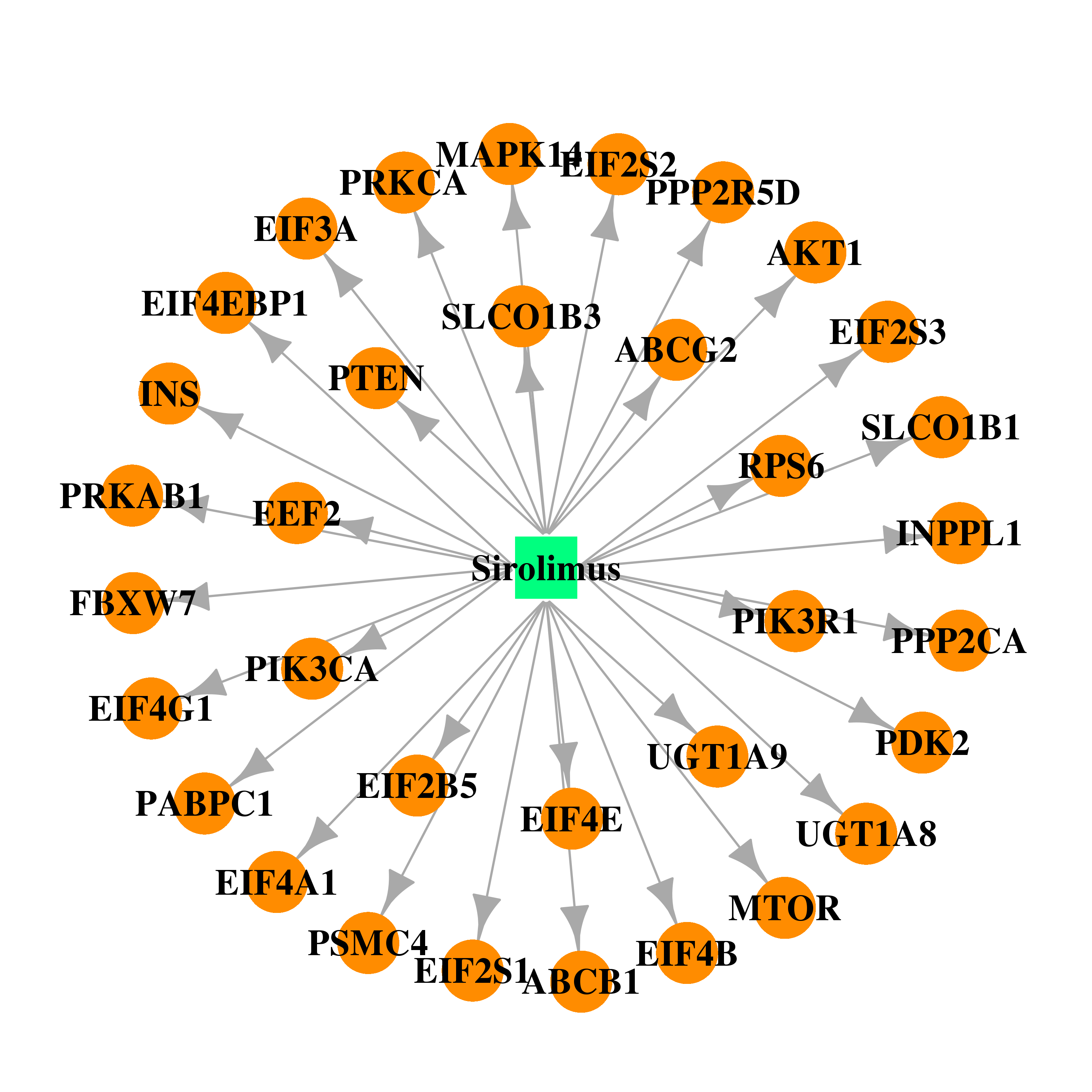 |  |
| Top |
| Cross referenced IDs for EIF4G1 |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |