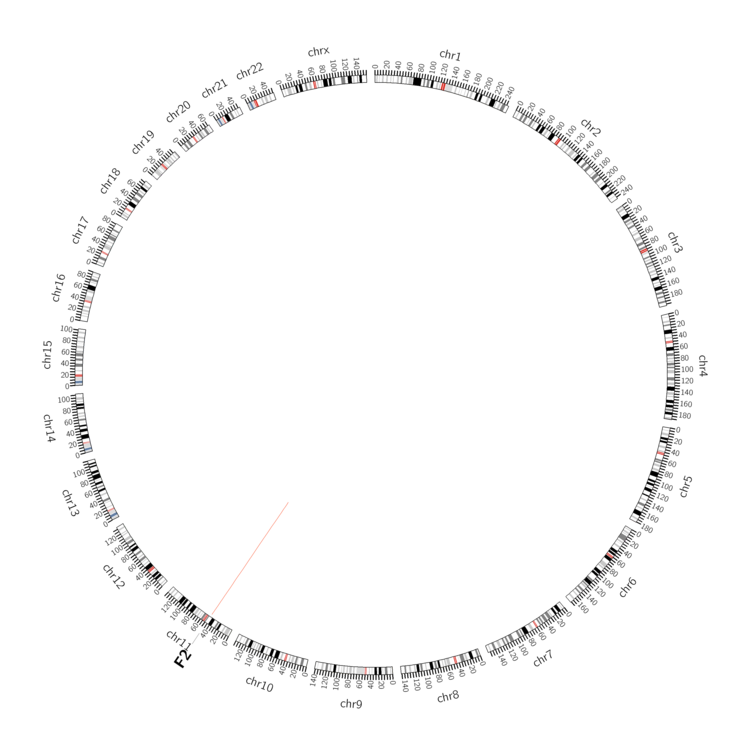
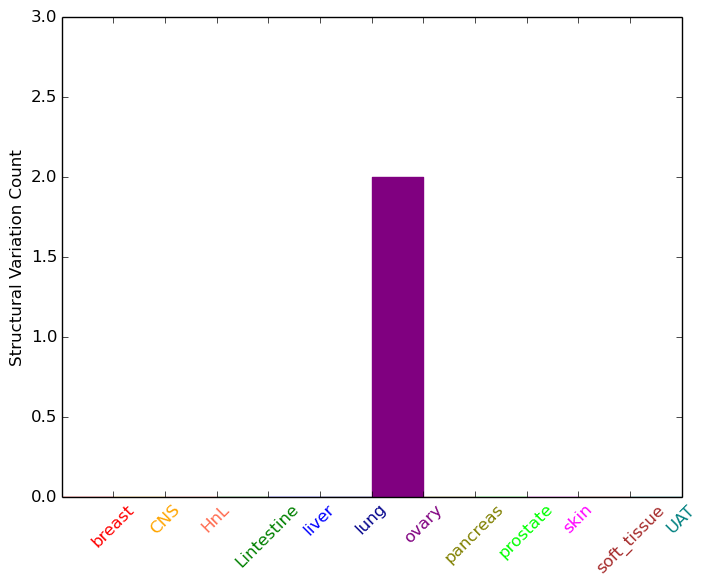
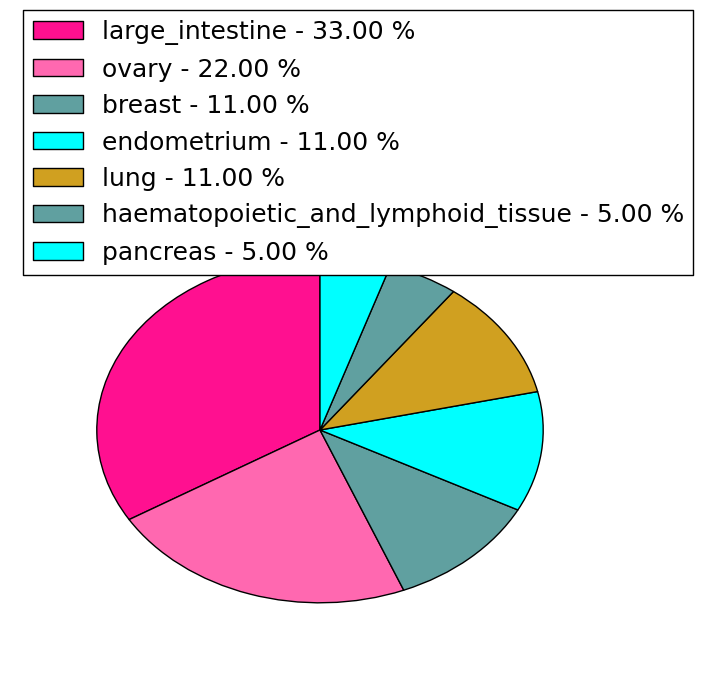
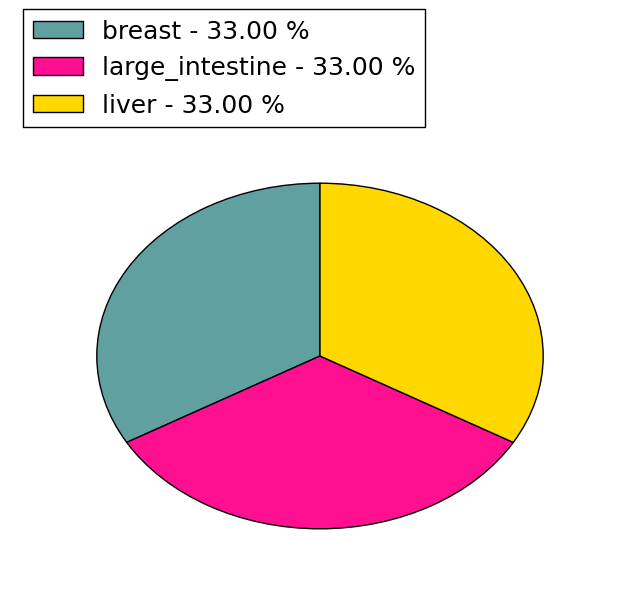
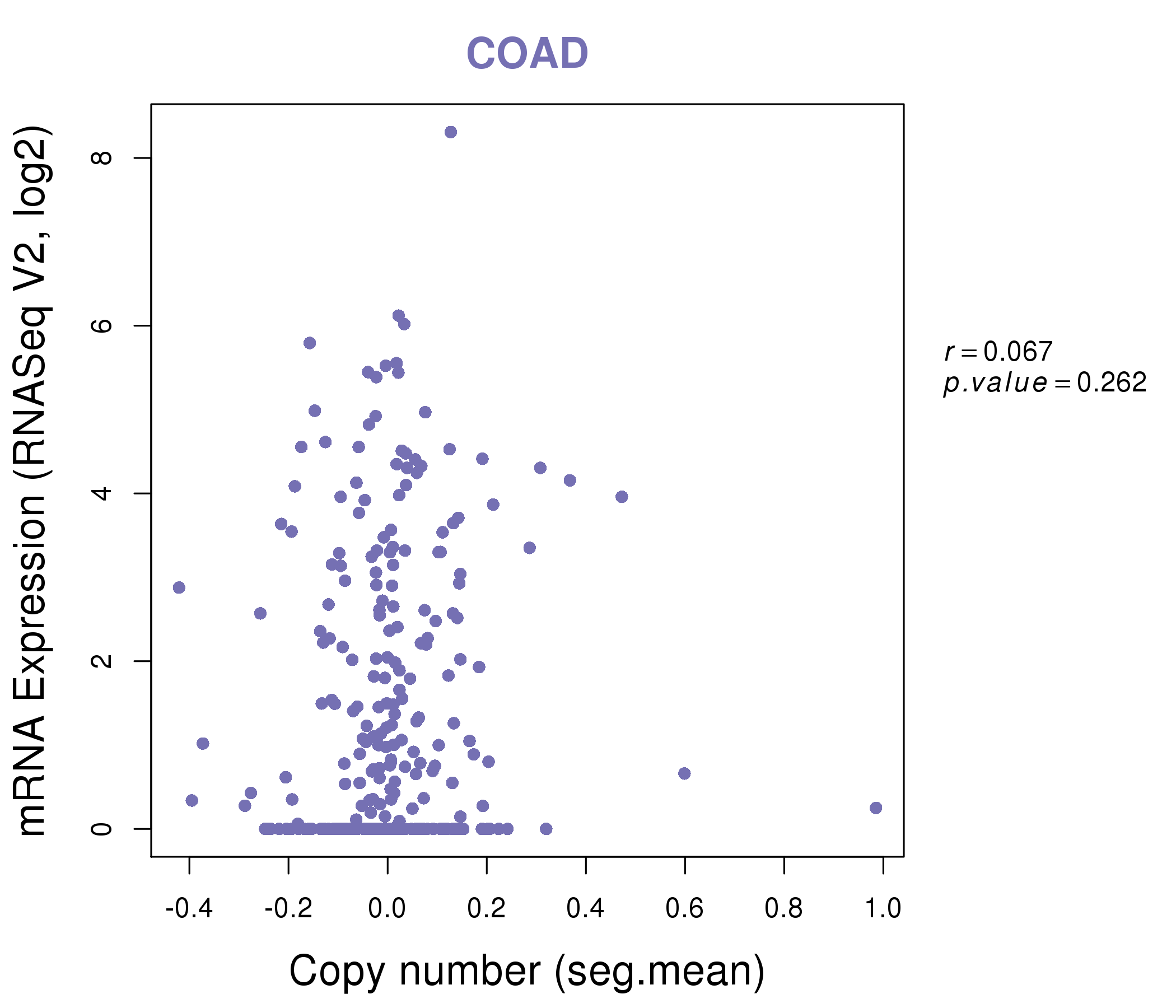
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB00170 | coagulation factor II (thrombin) | approved; nutraceutical | Menadione |  |  |
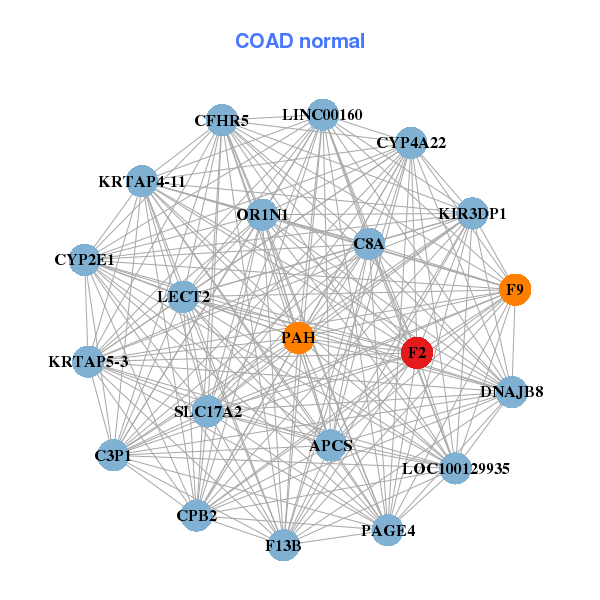
| DB00278 | coagulation factor II (thrombin) | approved; investigational | Argatroban | 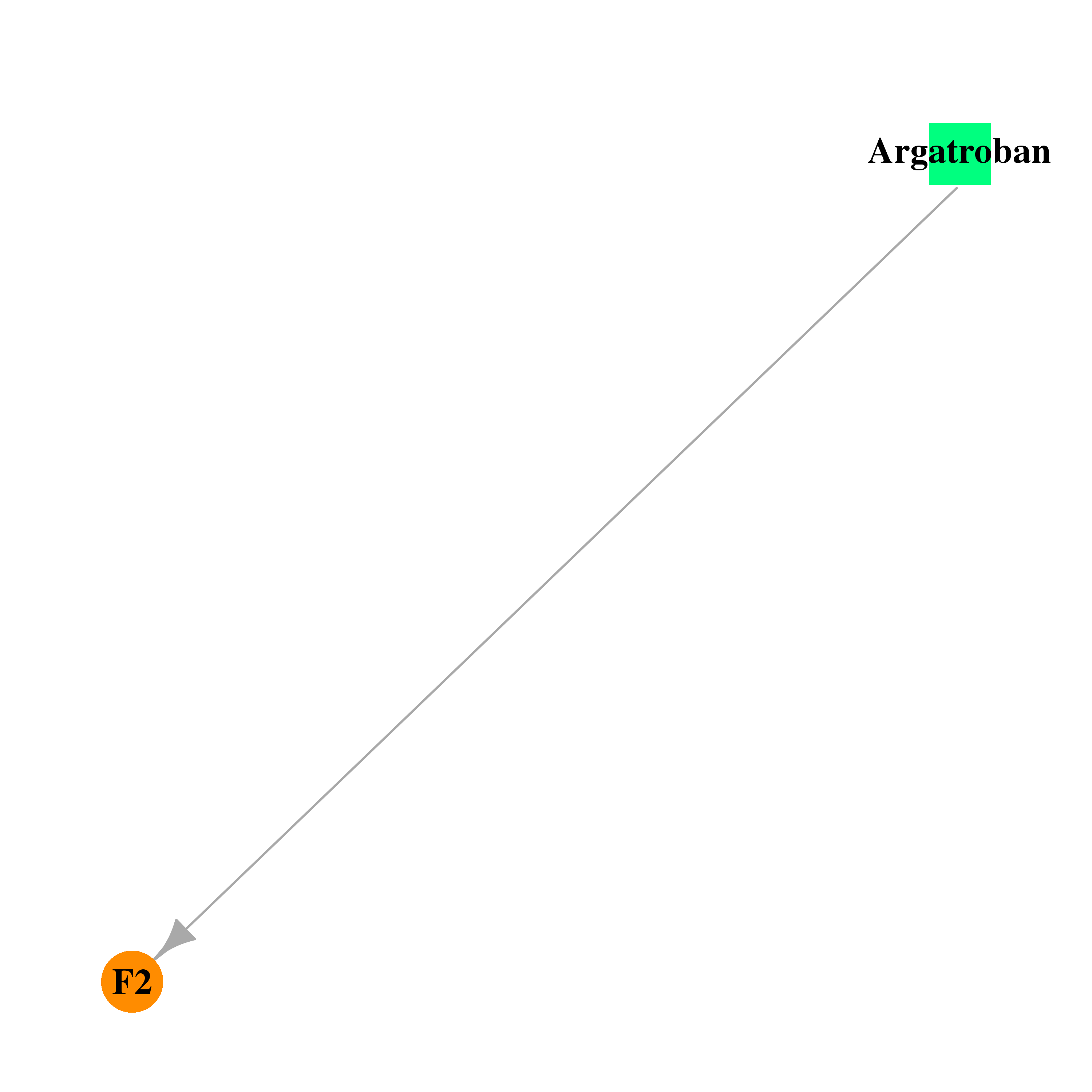 | 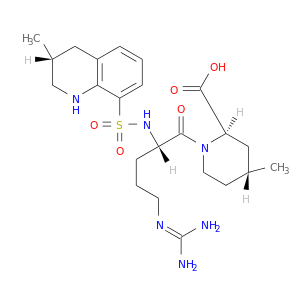 |
| DB01123 | coagulation factor II (thrombin) | approved | Proflavine |  | 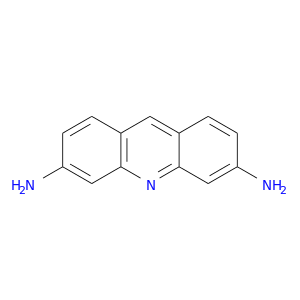 |
| DB01766 | coagulation factor II (thrombin) | experimental | Beta-(2-Naphthyl)-Alanine |  | 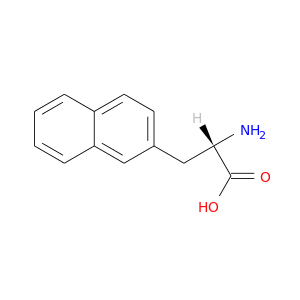 |
| DB01767 | coagulation factor II (thrombin) | experimental | Hemi-Babim |  |  |
| DB02193 | coagulation factor II (thrombin) | experimental | 2-(2-Hydroxy-Phenyl)-1h-Benzoimidazole-5-Carboxamidine |  |  |
| DB02351 | coagulation factor II (thrombin) | experimental | Hirulog |  |  |
| DB02723 | coagulation factor II (thrombin) | experimental | 4-Oxo-2-Phenylmethanesulfonyl-Octahydro-Pyrrolo[1,2-a]Pyrazine-6-Carboxylic Acid [1-(N-Hydroxycarbamimidoyl)-Piperidin-4-Ylmethyl]-Amide |  |  |
| DB03136 | coagulation factor II (thrombin) | experimental | 4-Iodobenzo[B]Thiophene-2-Carboxamidine |  |  |
| DB03159 | coagulation factor II (thrombin) | experimental | CRA_8696 |  |  |
| DB03847 | coagulation factor II (thrombin) | experimental | Gamma-Carboxy-Glutamic Acid |  |  |
| DB03865 | coagulation factor II (thrombin) | experimental | 6-Chloro-2-(2-Hydroxy-Biphenyl-3-Yl)-1h-Indole-5-Carboxamidine |  |  |
| DB04136 | coagulation factor II (thrombin) | experimental | Lysophosphotidylserine |  | 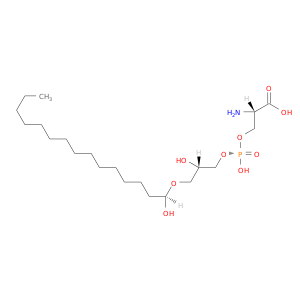 |
| DB04591 | coagulation factor II (thrombin) | experimental | N-{2,2-DIFLUORO-2-[(2R)-PIPERIDIN-2-YL]ETHYL}-2-[2-(1H-1,2,4-TRIAZOL-1-YL)BENZYL][1,3]OXAZOLO[4,5-C]PYRIDIN-4-AMINE |  |  |
| DB04697 | coagulation factor II (thrombin) | experimental | TRANS-4-(GUANIDINOMETHYL)-CYCLOHEXANE-L-YL-D-3-CYCLOHEXYLALANYL-L-AZETIDINE-2-YL-D-TYROSINYL-L-HOMOARGININAMIDE |  | 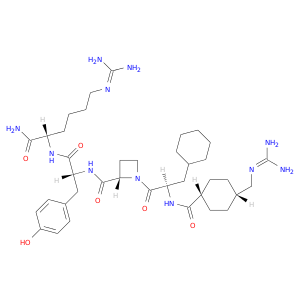 |
| DB04722 | coagulation factor II (thrombin) | experimental | 2-(3-CHLORO-6-{[2,2-DIFLUORO-2-(1-OXIDOPYRIDIN-2-YL)ETHYL]AMINO}-1-OXIDOPYRIDIN-2-YL)-N-[1-(3-CHLOROPHENYL)ETHYL]ACETAMIDE |  | 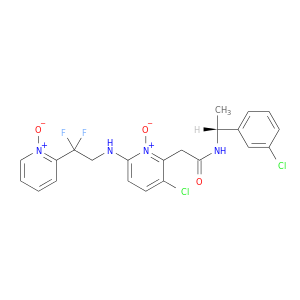 |
| DB04771 | coagulation factor II (thrombin) | experimental | 1-GUANIDINO-4-(N-NITRO-BENZOYLAMINO-L-LEUCYL-L-PROLYLAMINO)BUTANE |  | 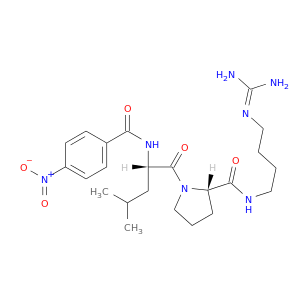 |
| DB04772 | coagulation factor II (thrombin) | experimental | 1-GUANIDINO-4-(N-PHENYLMETHANESULFONYL-L-LEUCYL-L-PROLYLAMINO)BUTANE |  |  |
| DB04786 | coagulation factor II (thrombin) | approved | Suramin |  |  |
| DB04898 | coagulation factor II (thrombin) | approved; withdrawn; investigational | Ximelagatran |  |  |
| DB06695 | coagulation factor II (thrombin) | approved | Dabigatran etexilate |  |  |
| DB06838 | coagulation factor II (thrombin) | experimental | methyl L-phenylalaninate |  | 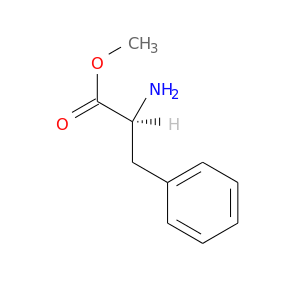 |
| DB06841 | coagulation factor II (thrombin) | experimental | D-phenylalanyl-N-[(1S)-4-{[amino(iminio)methyl]amino}-1-(chloroacetyl)butyl]-L-prolinamide |  | 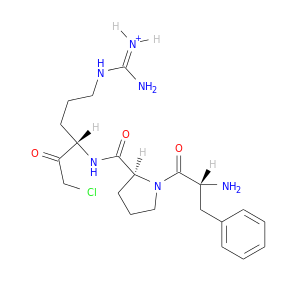 |
| DB06845 | coagulation factor II (thrombin) | experimental | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentylamino)ethanoyl)pyrrolidine-2-carboxamide |  | 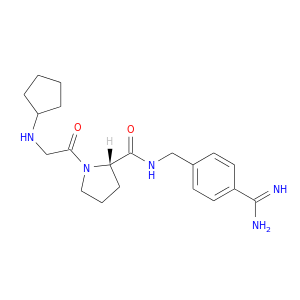 |
| DB06850 | coagulation factor II (thrombin) | experimental | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexylamino)ethanoyl)pyrrolidine-2-carboxamide |  | 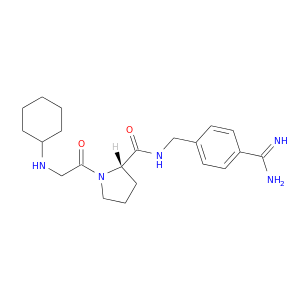 |
| DB06853 | coagulation factor II (thrombin) | experimental | N-cycloheptylglycyl-N-(4-carbamimidoylbenzyl)-L-prolinamide |  | 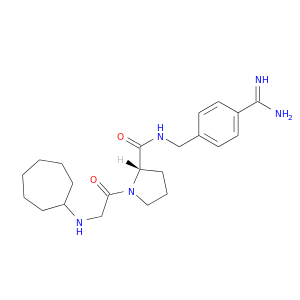 |
| DB06854 | coagulation factor II (thrombin) | experimental | 2-(2-HYDROXY-BIPHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE |  | 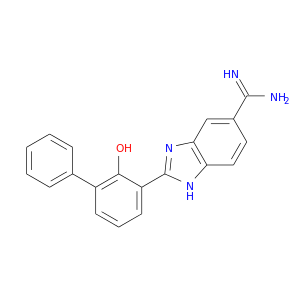 |
| DB06858 | coagulation factor II (thrombin) | experimental | N-cyclooctylglycyl-N-(4-carbamimidoylbenzyl)-L-prolinamide |  | 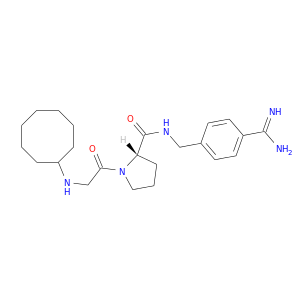 |
| DB06859 | coagulation factor II (thrombin) | experimental | N-ALLYL-5-AMIDINOAMINOOXY-PROPYLOXY-3-CHLORO-N-CYCLOPENTYLBENZAMIDE |  | 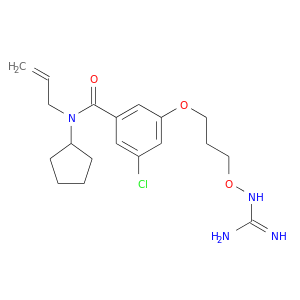 |
| DB06861 | coagulation factor II (thrombin) | experimental | 6-(2-HYDROXY-CYCLOPENTYL)-7-OXO-HEPTANAMIDINE |  | 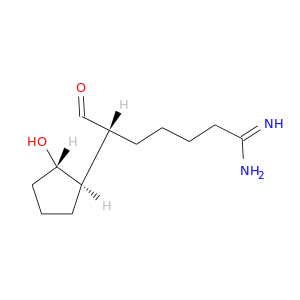 |
| DB06865 | coagulation factor II (thrombin) | experimental | 6-CARBAMIMIDOYL-2-[2-HYDROXY-6-(4-HYDROXY-PHENYL)-INDAN-1-YL]-HEXANOIC ACID |  |  |
| DB06866 | coagulation factor II (thrombin) | experimental | 6-CARBAMIMIDOYL-2-[2-HYDROXY-5-(3-METHOXY-PHENYL)-INDAN-1-YL]-HEXANOIC ACID |  | 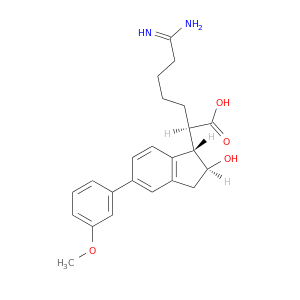 |
| DB06868 | coagulation factor II (thrombin) | experimental | N-(3-chlorobenzyl)-1-(4-methylpentanoyl)-L-prolinamide |  | 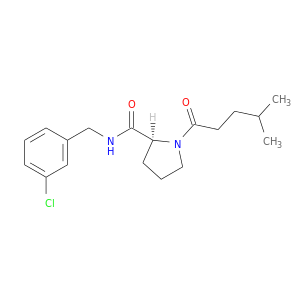 |
| DB06869 | coagulation factor II (thrombin) | experimental | 1-[2-AMINO-2-CYCLOHEXYL-ACETYL]-PYRROLIDINE-3-CARBOXYLIC ACID 5-CHLORO-2-(2-ETHYLCARBAMOYL-ETHOXY)-BENZYLAMIDE |  | 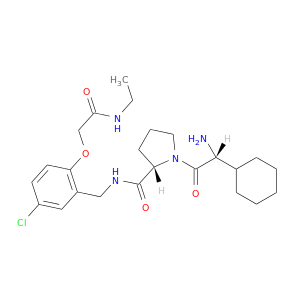 |
| DB06878 | coagulation factor II (thrombin) | experimental | 1-[(2R)-2-aminobutanoyl]-N-(3-chlorobenzyl)-L-prolinamide |  |  |
| DB06911 | coagulation factor II (thrombin) | experimental | D-leucyl-N-(3-chlorobenzyl)-L-prolinamide |  | 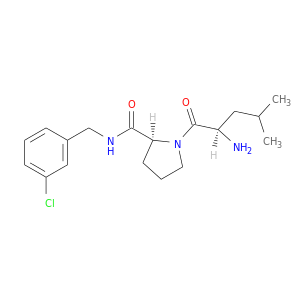 |
| DB06919 | coagulation factor II (thrombin) | experimental | D-phenylalanyl-N-(3-chlorobenzyl)-L-prolinamide |  | 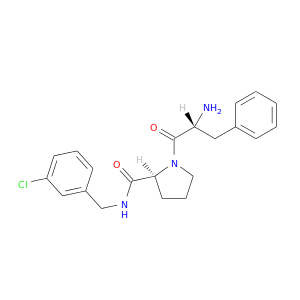 |
| DB06929 | coagulation factor II (thrombin) | experimental | 1-butanoyl-N-(4-carbamimidoylbenzyl)-L-prolinamide |  | 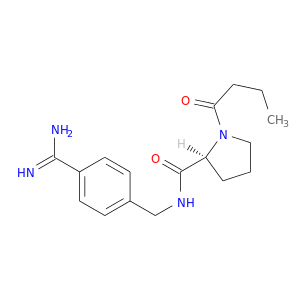 |
| DB06936 | coagulation factor II (thrombin) | experimental | N-(4-carbamimidoylbenzyl)-1-(4-methylpentanoyl)-L-prolinamide |  | 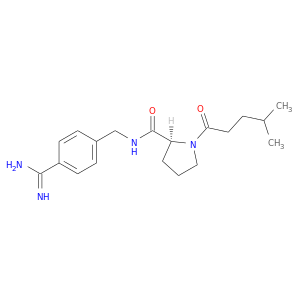 |
| DB06942 | coagulation factor II (thrombin) | experimental | N-(4-carbamimidoylbenzyl)-1-(3-phenylpropanoyl)-L-prolinamide |  | 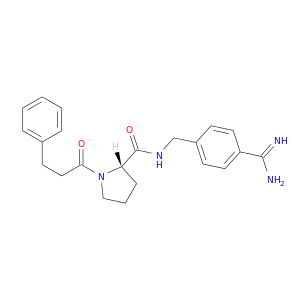 |
| DB06947 | coagulation factor II (thrombin) | experimental | 1-[(2R)-2-aminobutanoyl]-N-(4-carbamimidoylbenzyl)-L-prolinamide |  | 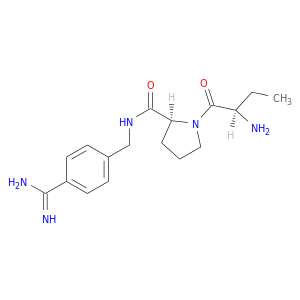 |
| DB06996 | coagulation factor II (thrombin) | experimental | D-leucyl-N-(4-carbamimidoylbenzyl)-L-prolinamide |  | 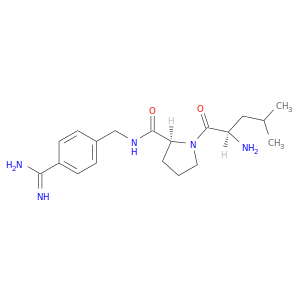 |
| DB07005 | coagulation factor II (thrombin) | experimental | D-phenylalanyl-N-{4-[amino(iminio)methyl]benzyl}-L-prolinamide |  | 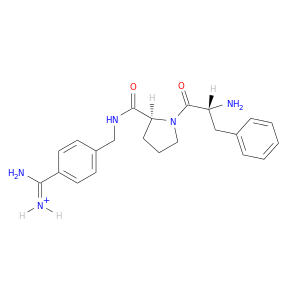 |
| DB07016 | coagulation factor II (thrombin) | experimental | (3R)-8-(dioxidosulfanyl)-3-methyl-1,2,3,4-tetrahydroquinoline |  | 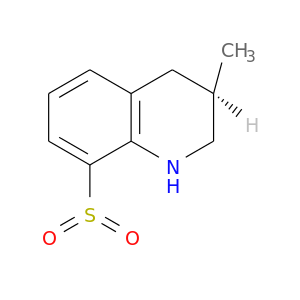 |
| DB07027 | coagulation factor II (thrombin) | experimental | D-phenylalanyl-N-(3-fluorobenzyl)-L-prolinamide |  | 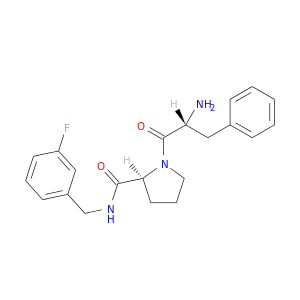 |
| DB07083 | coagulation factor II (thrombin) | experimental | beta-phenyl-D-phenylalanyl-N-propyl-L-prolinamide |  |  |
| DB07088 | coagulation factor II (thrombin) | experimental | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentyloxy)ethanoyl)pyrrolidine-2-carboxamide |  |  |
| DB07091 | coagulation factor II (thrombin) | experimental | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexyloxy)ethanoyl)pyrrolidine-2-carboxamide | 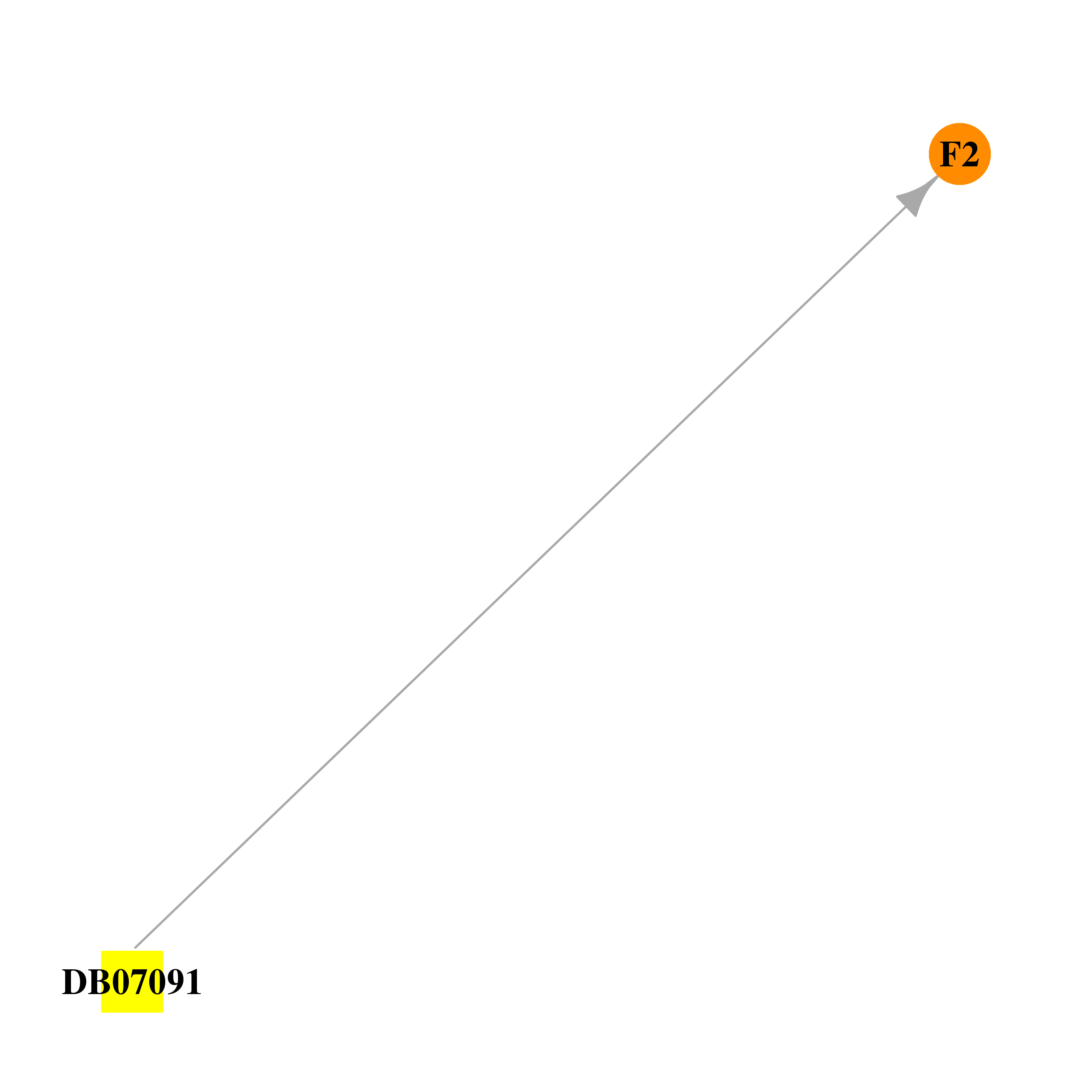 | 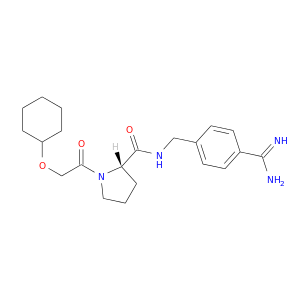 |
| DB07095 | coagulation factor II (thrombin) | experimental | (S)-N-(4-carbamimidoylbenzyl)-1-(3-cyclopentylpropanoyl)pyrrolidine-2-carboxamide |  |  |
| DB07105 | coagulation factor II (thrombin) | experimental | 2-[2-(4-CHLORO-PHENYLSULFANYL)-ACETYLAMINO]-3-(4-GUANIDINO-PHENYL)-PROPIONAMIDE |  |  |
| DB07120 | coagulation factor II (thrombin) | experimental | N4-(N,N-DIPHENYLCARBAMOYL)-AMINOGUANIDINE |  | 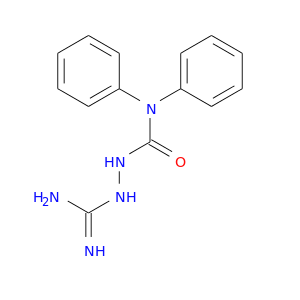 |
| DB07128 | coagulation factor II (thrombin) | experimental | N7-BUTYL-N2-(5-CHLORO-2-METHYLPHENYL)-5-METHYL[1,2,4]TRIAZOLO[1,5-A]PYRIMIDINE-2,7-DIAMINE |  | 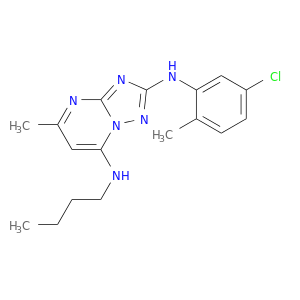 |
| DB07131 | coagulation factor II (thrombin) | experimental | (S)-N-(4-carbamimidoylbenzyl)-1-(3-cyclohexylpropanoyl)pyrrolidine-2-carboxamide |  | 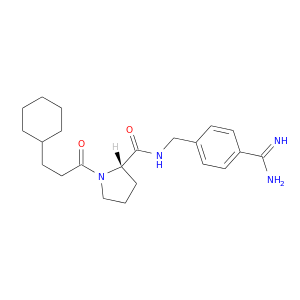 |
| DB07133 | coagulation factor II (thrombin) | experimental | D-phenylalanyl-N-(3-methylbenzyl)-L-prolinamide |  | 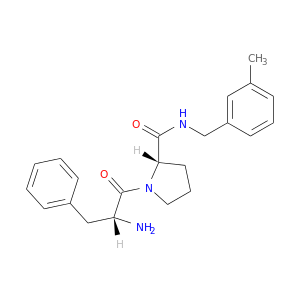 |
| DB07143 | coagulation factor II (thrombin) | experimental | D-phenylalanyl-N-benzyl-L-prolinamide |  | 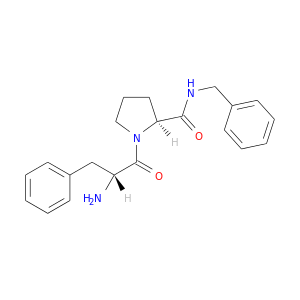 |
| DB07165 | coagulation factor II (thrombin) | experimental | N-(5-CHLORO-BENZO[B]THIOPHEN-3-YLMETHYL)-2-[6-CHLORO-OXO-3-(2-PYRIDIN-2-YL-ETHYLAMINO)-2H-PYRAZIN-1-YL]-ACETAMIDE |  | 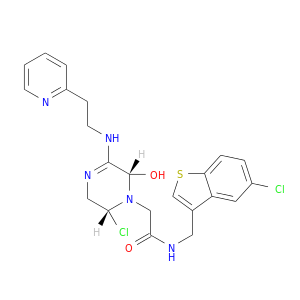 |
| DB07190 | coagulation factor II (thrombin) | experimental | 3-cyclohexyl-D-alanyl-N-(3-chlorobenzyl)-L-prolinamide |  |  |
| DB07211 | coagulation factor II (thrombin) | experimental | (2R)-2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}PROPENE-1-SULFONAMIDE |  | 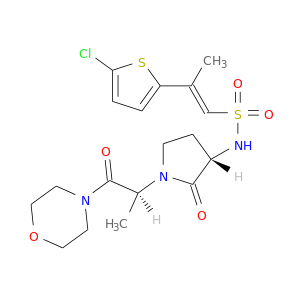 |
| DB07277 | coagulation factor II (thrombin) | experimental | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHANESULFONAMIDE |  | 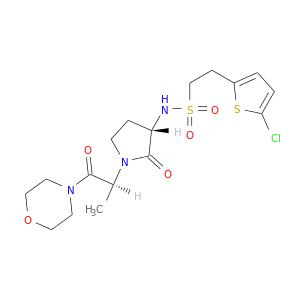 |
| DB07278 | coagulation factor II (thrombin) | experimental | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHENESULFONAMIDE |  | 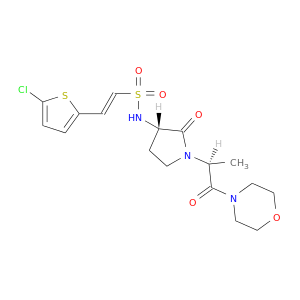 |
| DB07279 | coagulation factor II (thrombin) | experimental | N-ETHYL-N-ISOPROPYL-3-METHYL-5-{[(2S)-2-(PYRIDIN-4-YLAMINO)PROPYL]OXY}BENZAMIDE |  | 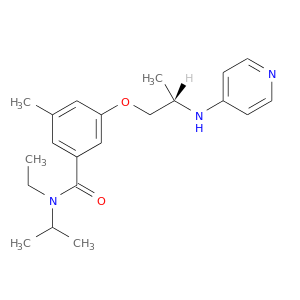 |
| DB07353 | coagulation factor II (thrombin) | experimental | 4-(2,5-DIAMINO-5-HYDROXY-PENTYL)-PHENOL |  | 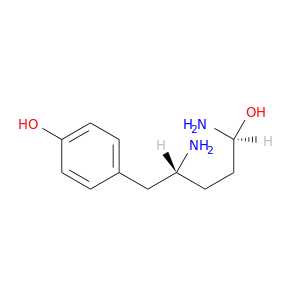 |
| DB07366 | coagulation factor II (thrombin) | experimental | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER |  | 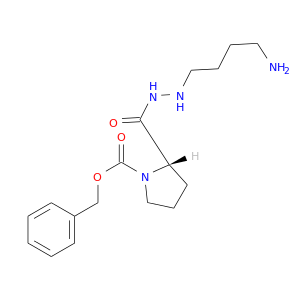 |
| DB07376 | coagulation factor II (thrombin) | experimental | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID) |  | 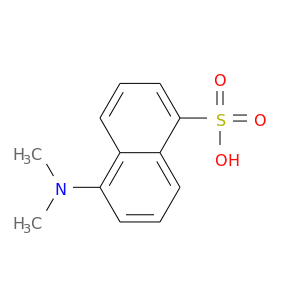 |
| DB07400 | coagulation factor II (thrombin) | experimental | 1-ETHOXYCARBONYL-D-PHE-PRO-2(4-AMINOBUTYL)HYDRAZINE |  | 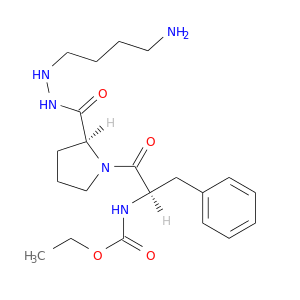 |
| DB07440 | coagulation factor II (thrombin) | experimental | 4-TERT-BUTYLBENZENESULFONIC ACID |  | 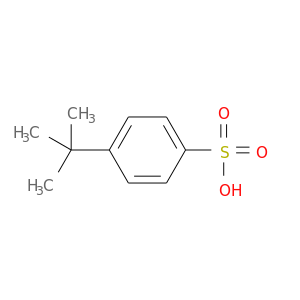 |
| DB07461 | coagulation factor II (thrombin) | experimental | 3-AMINO-3-BENZYL-9-CARBOXAMIDE[4.3.0]BICYCLO-1,6-DIAZANONAN-2-ONE |  | 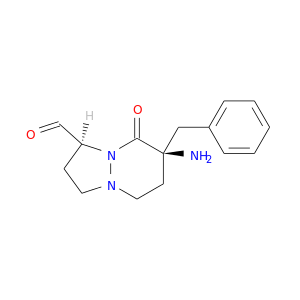 |
| DB07508 | coagulation factor II (thrombin) | experimental | 4-(5-BENZENESULFONYLAMINO-1-METHYL-1H-BENZOIMIDAZOL-2-YLMETHYL)-BENZAMIDINE |  |  |
| DB07515 | coagulation factor II (thrombin) | experimental | 1-(2-{[(6-AMINO-2-METHYLPYRIDIN-3-YL)METHYL]AMINO}ETHYL)-6-CHLORO-3-[(2,2-DIFLUORO-2-PYRIDIN-2-YLETHYL)AMINO]-1,4-DIHYDROPYRAZIN-2-OL |  |  |
| DB07521 | coagulation factor II (thrombin) | experimental | 6-CHLORO-1-(2-{[(5-CHLORO-1-BENZOTHIEN-3-YL)METHYL]AMINO}ETHYL)-3-[(2-PYRIDIN-2-YLETHYL)AMINO]-1,4-DIHYDROPYRAZIN-2-OL |  | 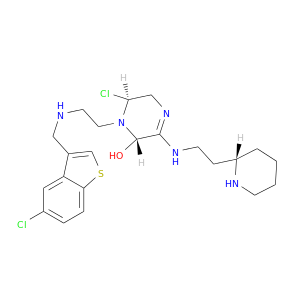 |
| DB07522 | coagulation factor II (thrombin) | experimental | N-[(2R,3S)-3-AMINO-2-HYDROXY-4-PHENYLBUTYL]NAPHTHALENE-2-SULFONAMIDE |  |  |
| DB07527 | coagulation factor II (thrombin) | experimental | N-[(2R,3S)-3-AMINO-2-HYDROXY-4-PHENYLBUTYL]-4-METHOXY-2,3,6-TRIMETHYLBENZENESULFONAMIDE |  | 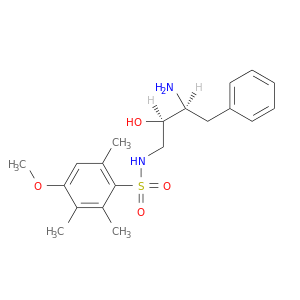 |
| DB07548 | coagulation factor II (thrombin) | experimental | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-6-PYRIDINYL)METHYL]ACETAMIDE |  |  |
| DB07549 | coagulation factor II (thrombin) | experimental | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-3-METHYL-6-PYRIDINYL)METHYL]ACETAMIDE |  | 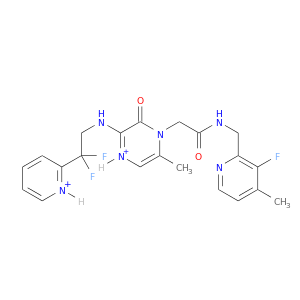 |
| DB07550 | coagulation factor II (thrombin) | experimental | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDO-2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUOROPHENYL)METHYL]ACETAMIDE |  | 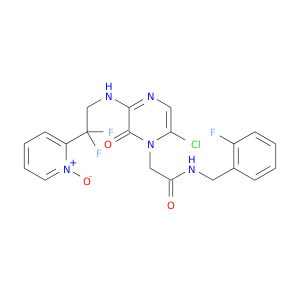 |
| DB07639 | coagulation factor II (thrombin) | experimental | 3-(7-DIAMINOMETHYL-NAPHTHALEN-2-YL)-PROPIONIC ACID ETHYL ESTER |  | 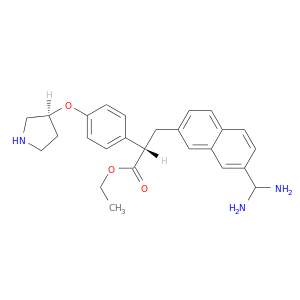 |
| DB07658 | coagulation factor II (thrombin) | experimental | AC-(D)PHE-PRO-BOROLYS-OH |  | 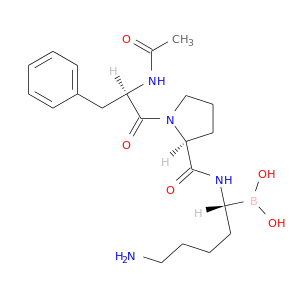 |
| DB07659 | coagulation factor II (thrombin) | experimental | AC-(D)PHE-PRO-BOROHOMOLYS-OH |  | 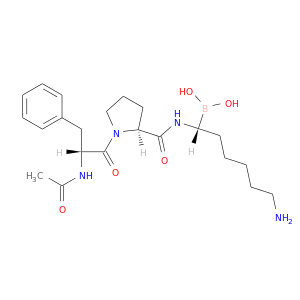 |
| DB07660 | coagulation factor II (thrombin) | experimental | AC-(D)PHE-PRO-BOROHOMOORNITHINE-OH |  | 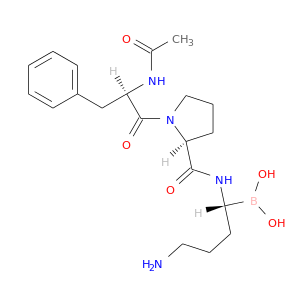 |
| DB07665 | coagulation factor II (thrombin) | experimental | N-[2-(carbamimidamidooxy)ethyl]-2-{6-cyano-3-[(2,2-difluoro-2-pyridin-2-ylethyl)amino]-2-fluorophenyl}acetamide |  |  |
| DB07718 | coagulation factor II (thrombin) | experimental | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID |  |  |
| DB07741 | coagulation factor II (thrombin) | experimental | 4-(1R,3AS,4R,8AS,8BR)-[1-DIFLUOROMETHYL-2-(4-FLUOROBENZYL)-3-OXODECAHYDROPYRROLO[3,4-A]PYRROLIZIN-4-YL]BENZAMIDINE |  | 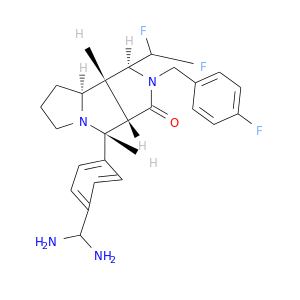 |
| DB07796 | coagulation factor II (thrombin) | experimental | (3ASR,4RS,8ASR,8BRS)-4-(2-(4-FLUOROBENZYL)-1,3-DIOXODEACAHYDROPYRROLO[3,4-A] PYRROLIZIN-4-YL)BENZAMIDINE |  | 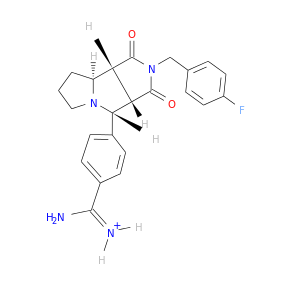 |
| DB07809 | coagulation factor II (thrombin) | experimental | 4-({[4-(3-METHYLBENZOYL)PYRIDIN-2-YL]AMINO}METHYL)BENZENECARBOXIMIDAMIDE |  |  |
| DB07897 | coagulation factor II (thrombin) | experimental | 1-(HYDROXYMETHYLENEAMINO)-8-HYDROXY-OCTANE |  |  |
| DB07934 | coagulation factor II (thrombin) | experimental | [[CYCLOHEXANESULFONYL-GLYCYL]-3[PYRIDIN-4-YL-AMINOMETHYL]ALANYL]PIPERIDINE |  | 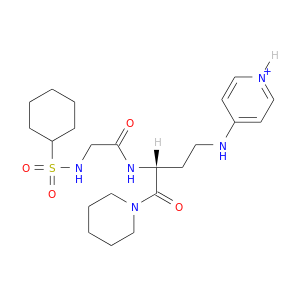 |
| DB07944 | coagulation factor II (thrombin) | experimental | N-{3-METHYL-5-[2-(PYRIDIN-4-YLAMINO)-ETHOXY]-PHENYL}-BENZENESULFONAMIDE |  | 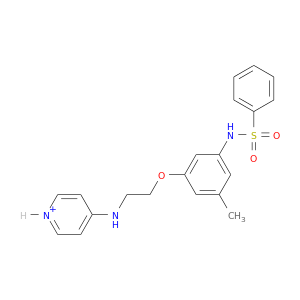 |
| DB07946 | coagulation factor II (thrombin) | experimental | N-[2-({[amino(imino)methyl]amino}oxy)ethyl]-2-{6-chloro-3-[(2,2-difluoro-2-phenylethyl)amino]-2-fluorophenyl}acetamide |  | 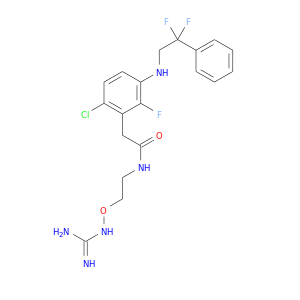 |
| DB08061 | coagulation factor II (thrombin) | experimental | 4-[3-(4-CHLOROPHENYL)-1H-PYRAZOL-5-YL]PIPERIDINE |  | 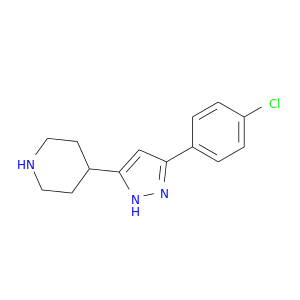 |
| DB08062 | coagulation factor II (thrombin) | experimental | 3-(4-CHLOROPHENYL)-5-(METHYLTHIO)-4H-1,2,4-TRIAZOLE |  | 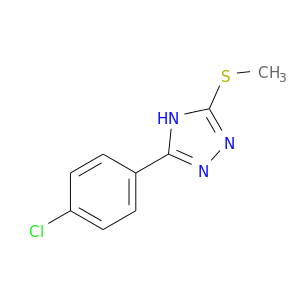 |
| DB08152 | coagulation factor II (thrombin) | experimental | {(2S)-1-[N-(tert-butoxycarbonyl)glycyl]pyrrolidin-2-yl}methyl (3-chlorophenyl)acetate |  | 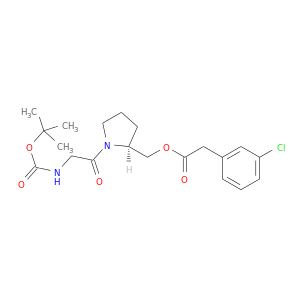 |
| DB08187 | coagulation factor II (thrombin) | experimental | METHYL-PHE-PRO-AMINO-CYCLOHEXYLGLYCINE |  | 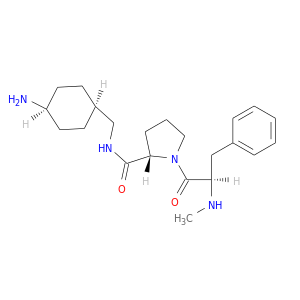 |
| DB08254 | coagulation factor II (thrombin) | experimental | 2-NAPHTHALENESULFONIC ACID |  | 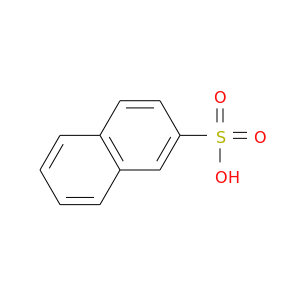 |
| DB08422 | coagulation factor II (thrombin) | experimental | [PHENYLALANINYL-PROLINYL]-[2-(PYRIDIN-4-YLAMINO)-ETHYL]-AMINE |  |  |
| DB08546 | coagulation factor II (thrombin) | experimental | 4-[(3AS,4R,7R,8AS,8BR)-2-(1,3-BENZODIOXOL-5-YLMETHYL)-7-HYDROXY-1,3-DIOXODECAHYDROPYRROLO[3,4-A]PYRROLIZIN-4-YL]BENZENECARBOXIMIDAMIDE |  |  |
| DB08624 | coagulation factor II (thrombin) | experimental | BENZOTHIAZOLE |  |  |
| DB00945 | coagulation factor II (thrombin) | approved | Acetylsalicylic acid | 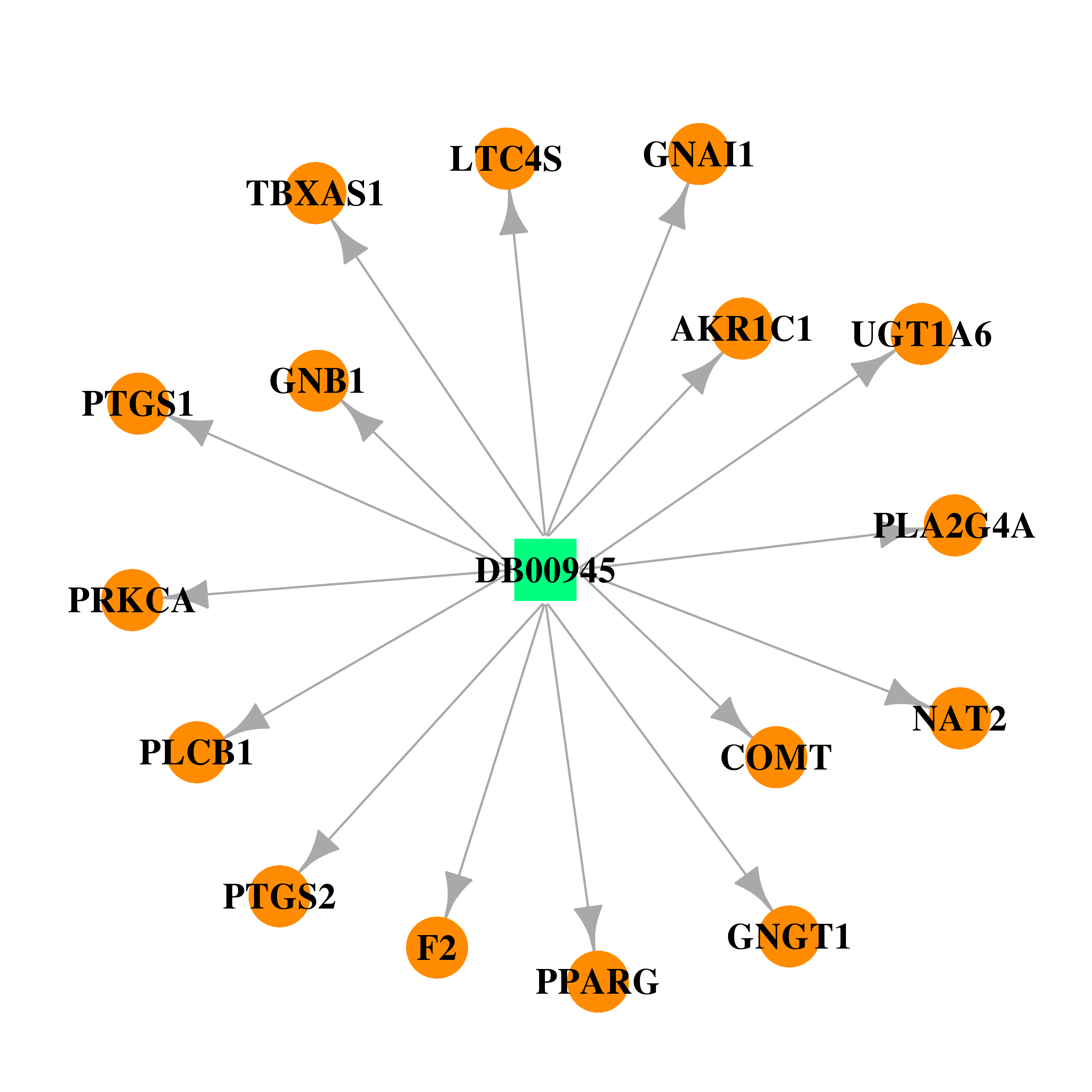 | 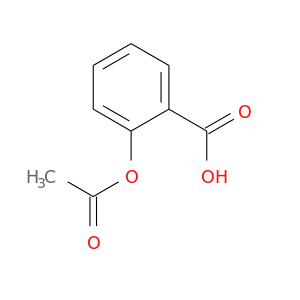 |
| DB00682 | coagulation factor II (thrombin) | approved | Warfarin |  | 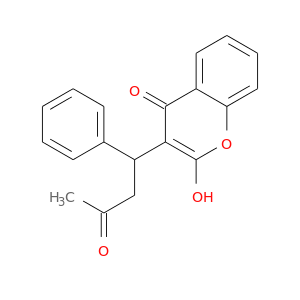 |
| DB00269 | coagulation factor II (thrombin) | approved | Chlorotrianisene | 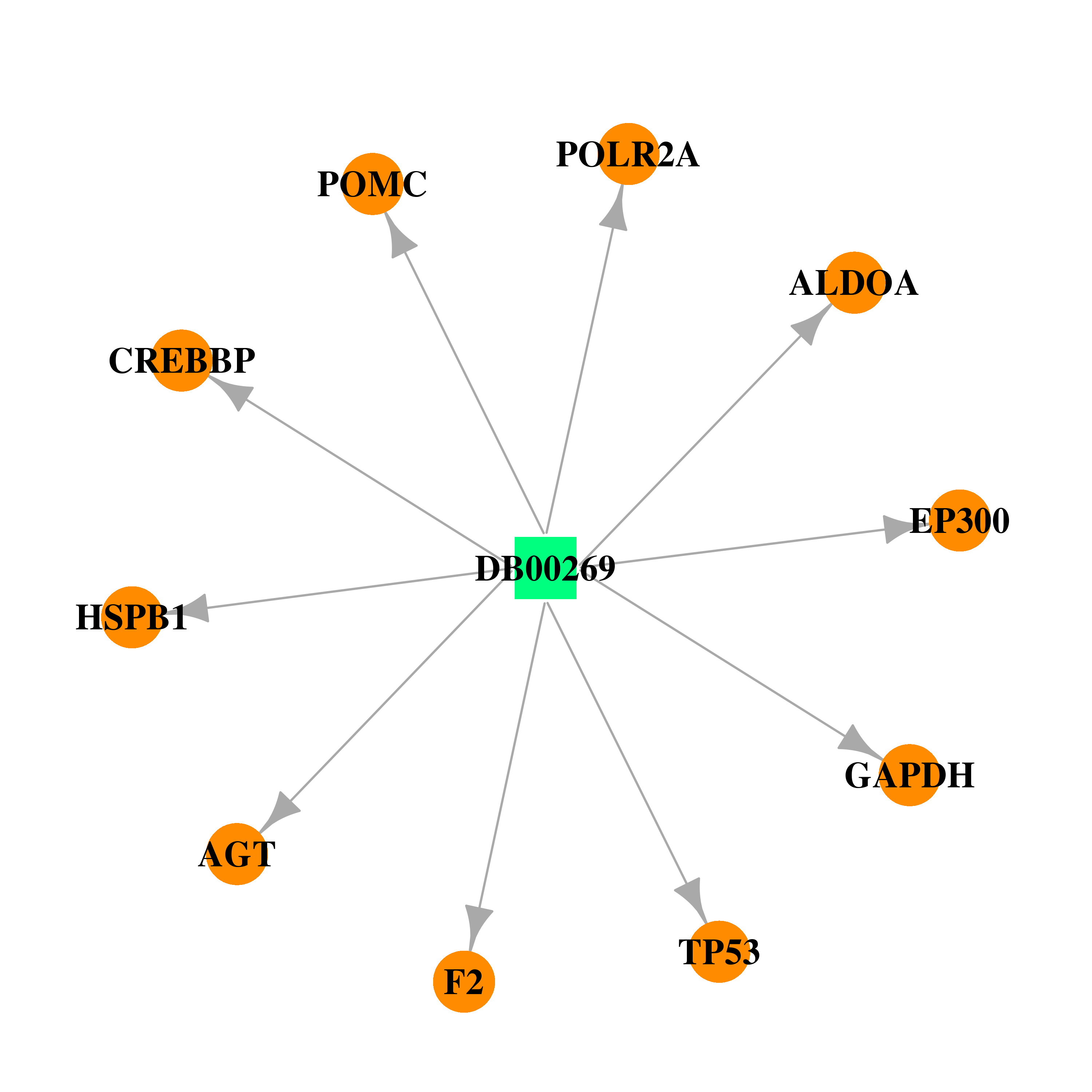 |  |
| DB00286 | coagulation factor II (thrombin) | approved | Conjugated Estrogens | 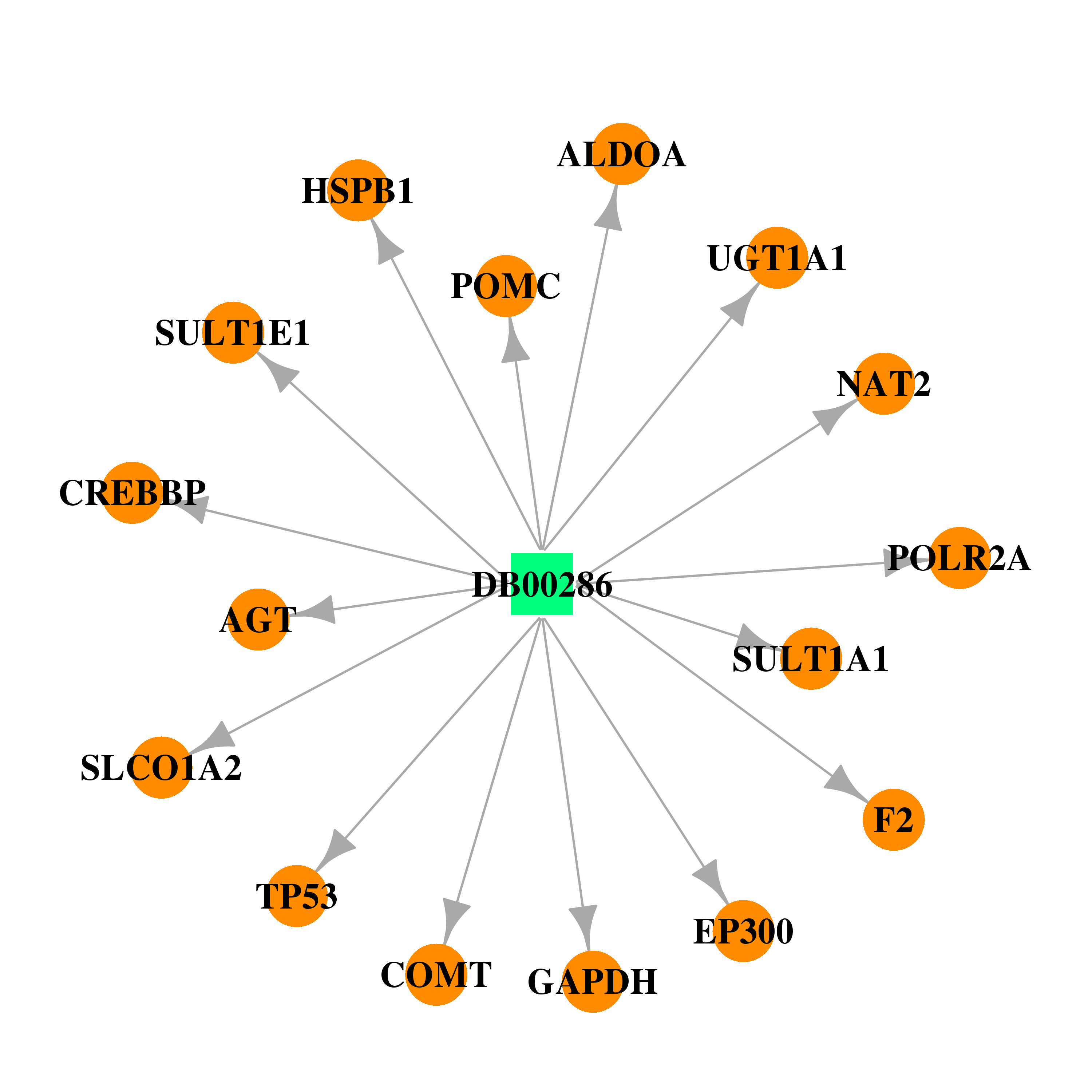 |  |
| DB00890 | coagulation factor II (thrombin) | approved | Dienestrol |  |  |
| DB00255 | coagulation factor II (thrombin) | approved | Diethylstilbestrol |  |  |
| DB00783 | coagulation factor II (thrombin) | approved; investigational | Estradiol |  | 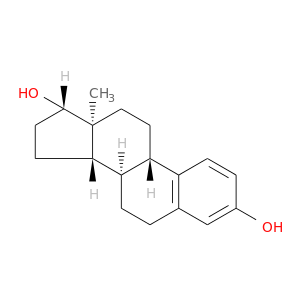 |
| DB00655 | coagulation factor II (thrombin) | approved | Estrone | 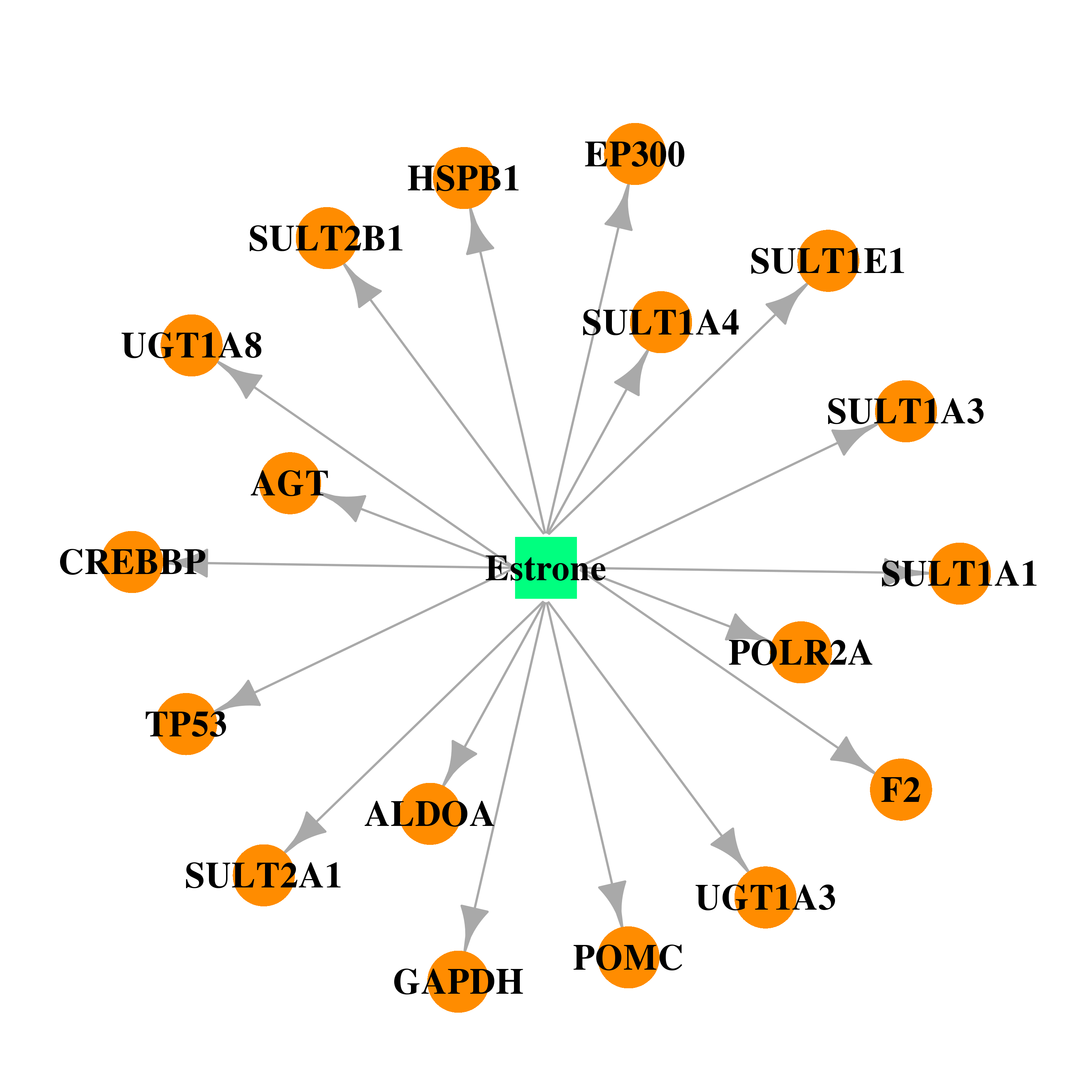 | 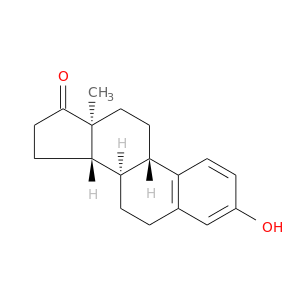 |
| DB00977 | coagulation factor II (thrombin) | approved | Ethinyl Estradiol | 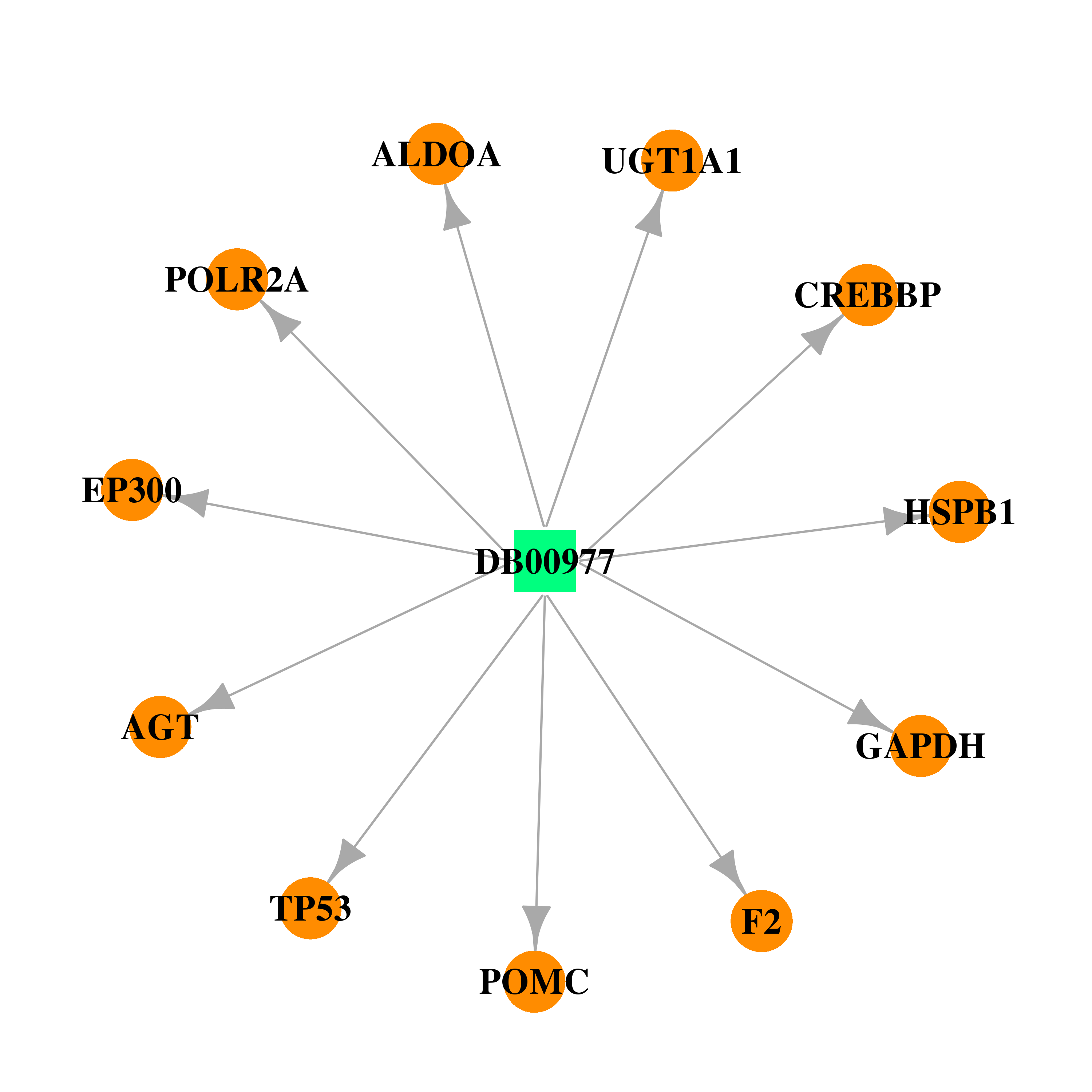 |  |
| DB00122 | coagulation factor II (thrombin) | approved; nutraceutical | Choline | 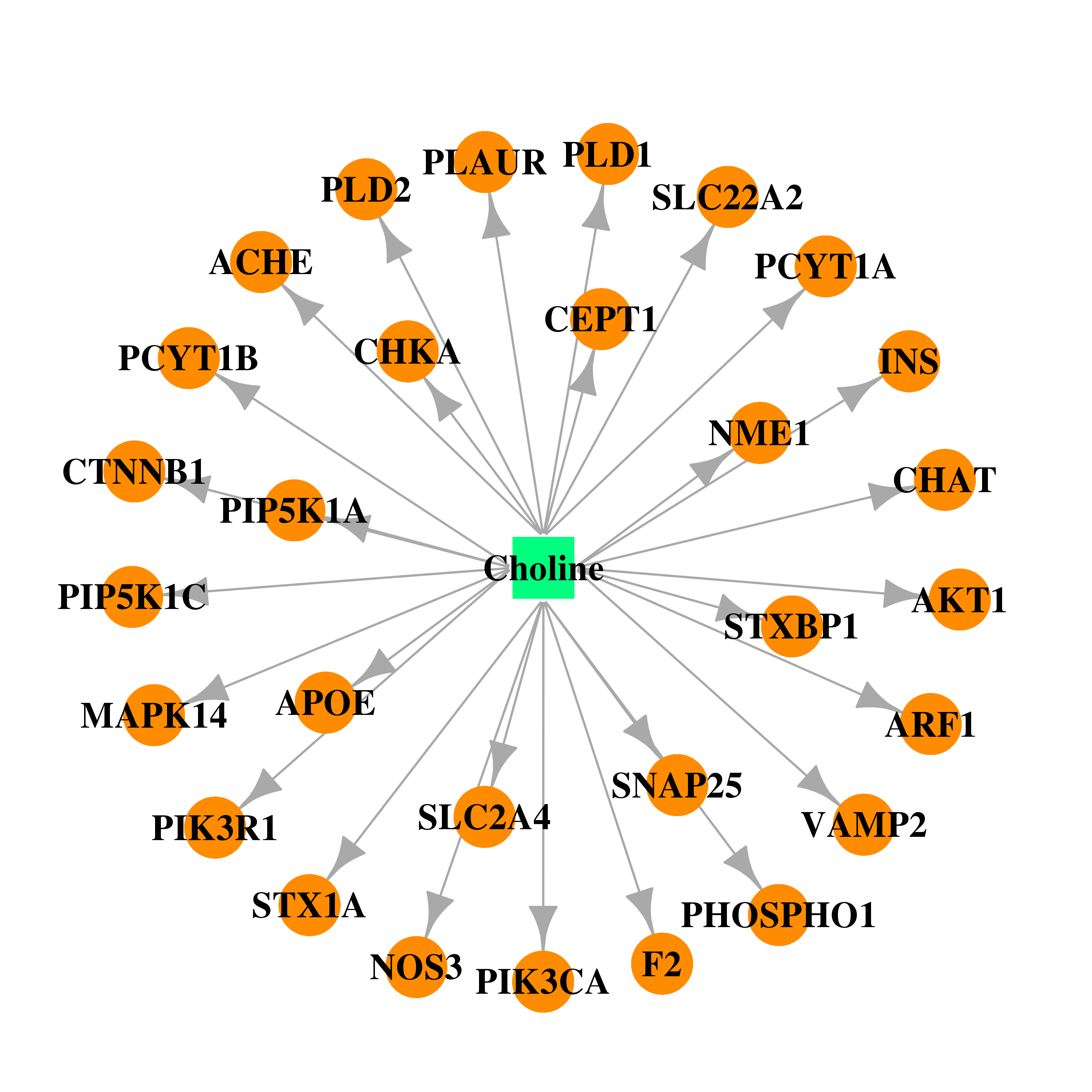 | 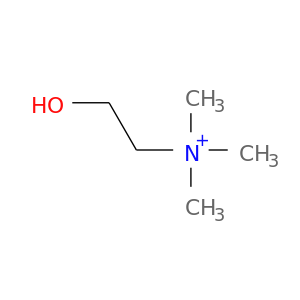 |
| DB00125 | coagulation factor II (thrombin) | approved; nutraceutical | L-Arginine | 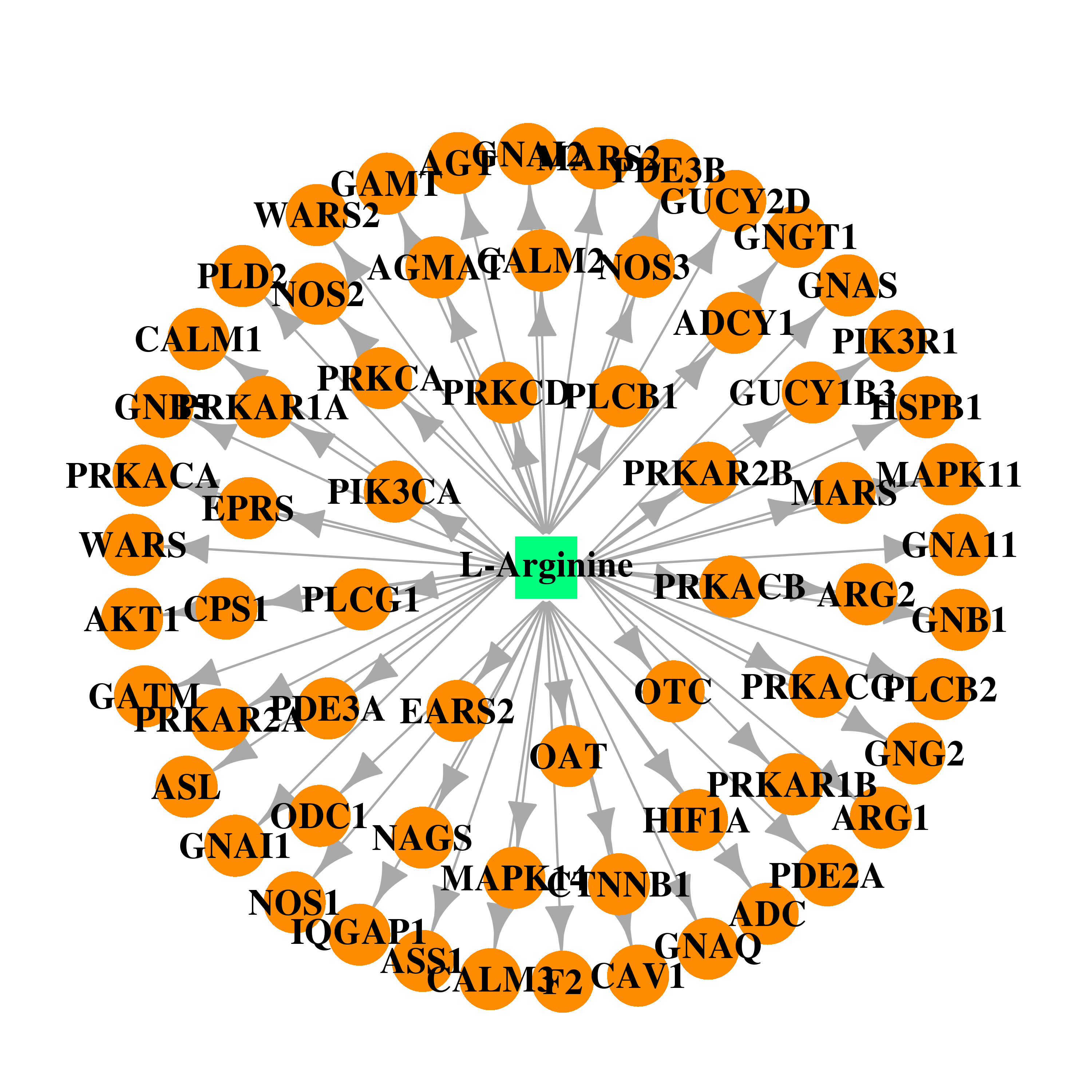 | 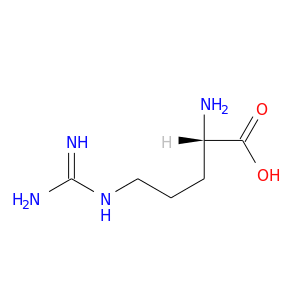 |
| DB00435 | coagulation factor II (thrombin) | approved | Nitric Oxide | 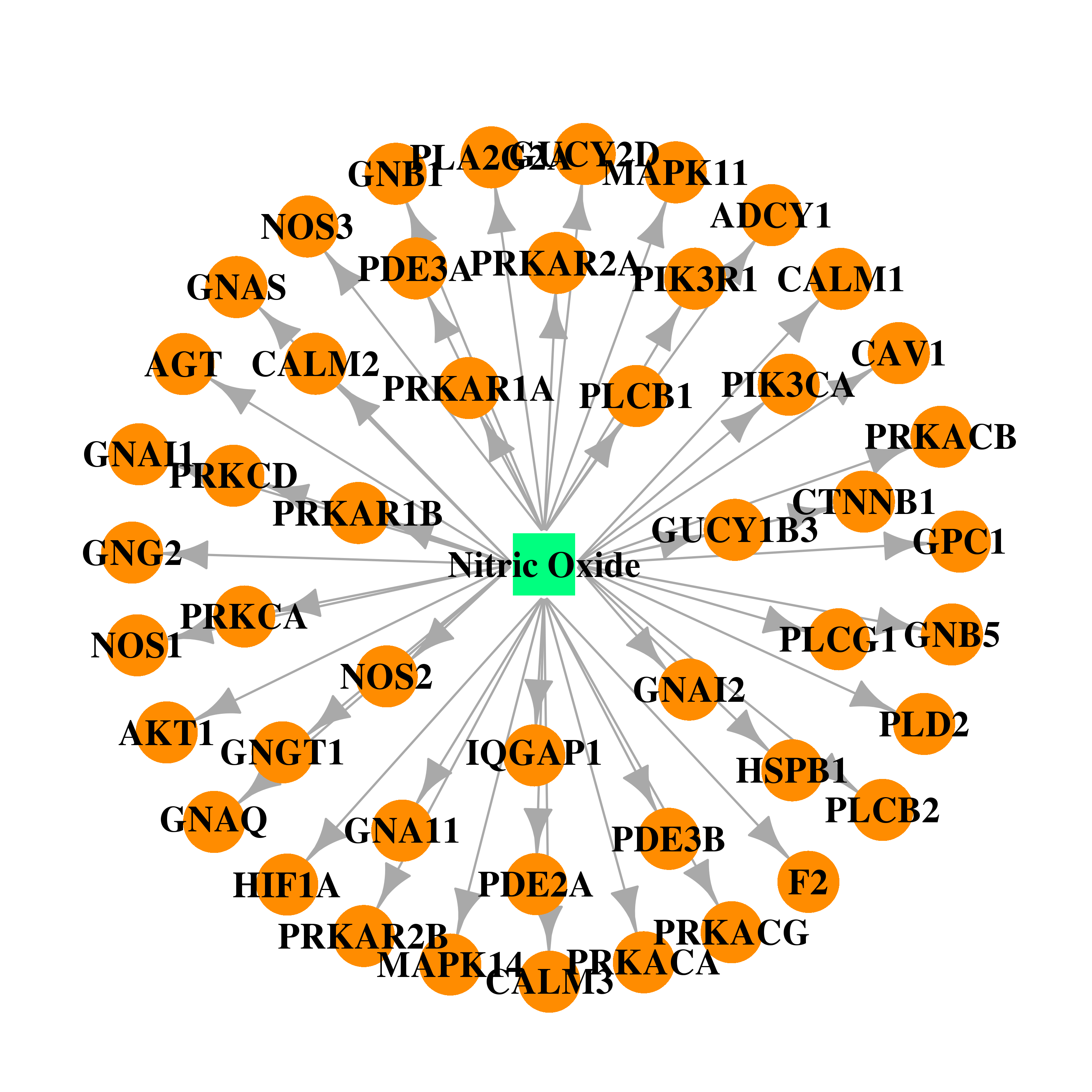 |  |
| DB00668 | coagulation factor II (thrombin) | approved | Epinephrine | 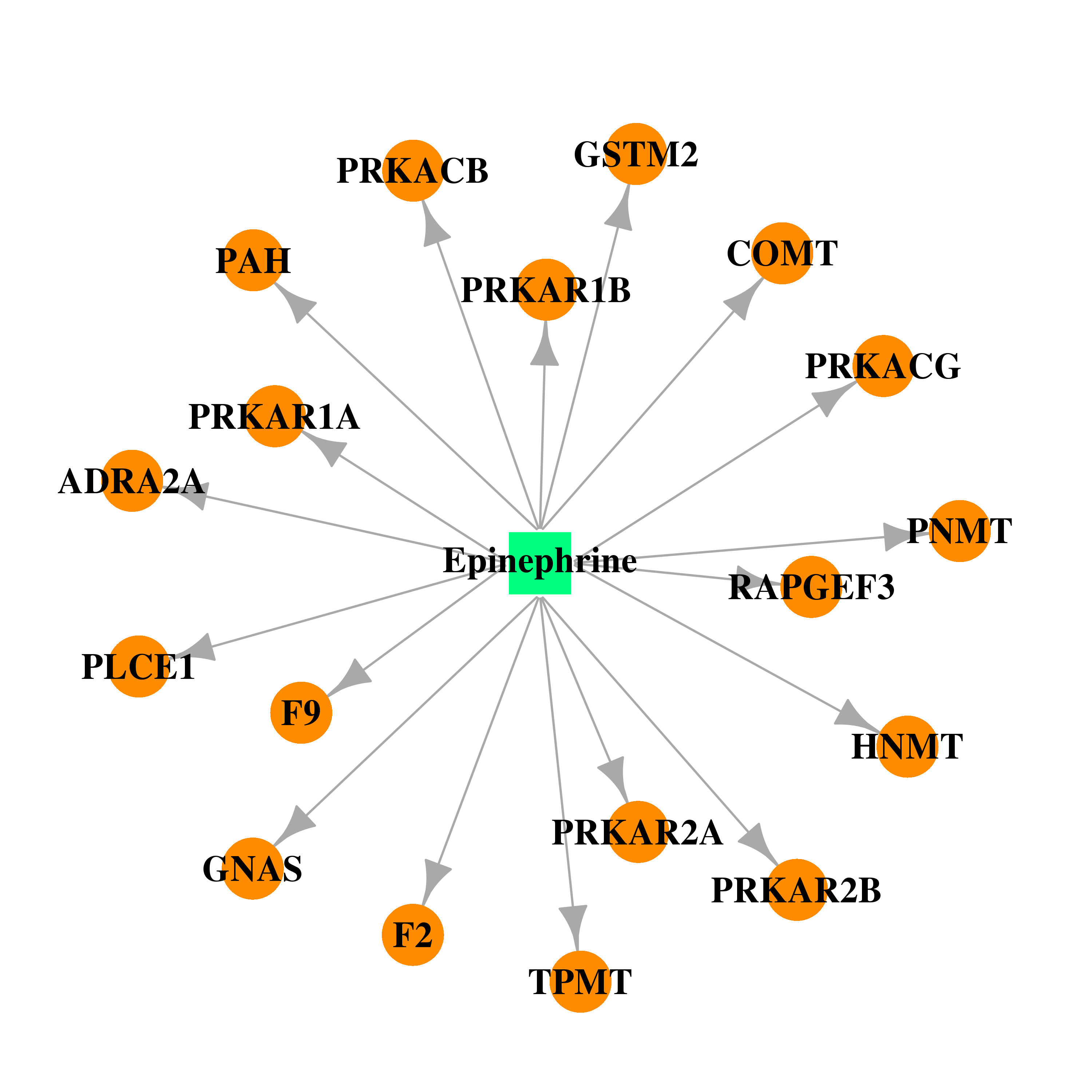 |  |
| DB00460 | coagulation factor II (thrombin) | approved; investigational | Verteporfin | 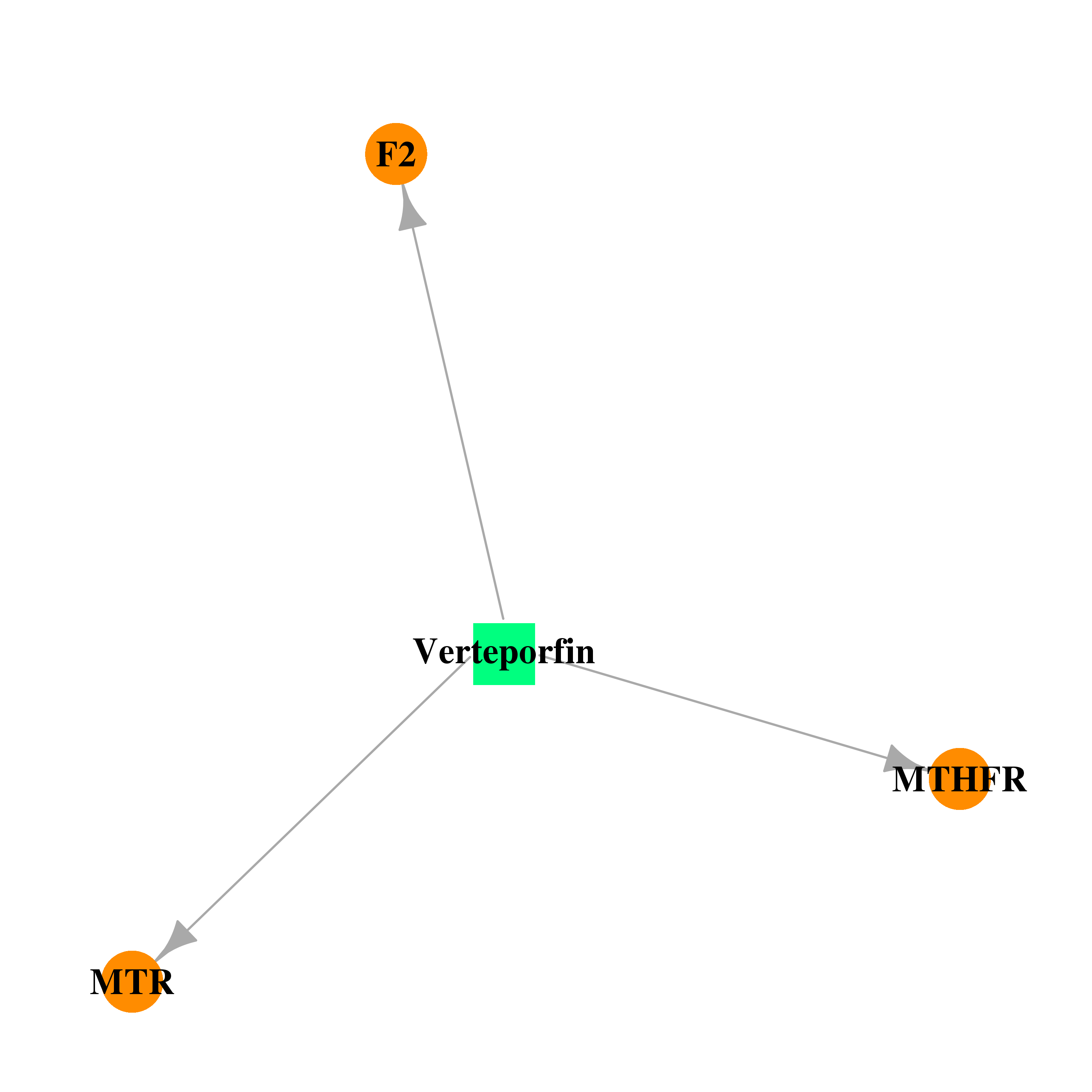 |  |
| DB00717 | coagulation factor II (thrombin) | approved | Norethindrone |  |  |