|
||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for FBXO4 |
| Top |
| Phenotypic Information for FBXO4(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: FBXO4 |
| Familial Cancer Database: FBXO4 | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
| . | ||||||||||||||
Oncogene 1 | Significant driver gene in | |||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| REACTOME_METABOLISM_OF_PROTEINS | |
| OMIM | 609090; gene. |
| Orphanet | |
| Disease | KEGG Disease: FBXO4 |
| MedGen: FBXO4 (Human Medical Genetics with Condition) | |
| ClinVar: FBXO4 | |
| Phenotype | MGI: FBXO4 (International Mouse Phenotyping Consortium) |
| PhenomicDB: FBXO4 | |
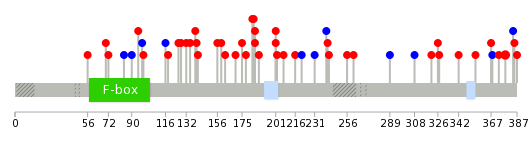
| Mutations for FBXO4 |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| There's no structural variation information in COSMIC data for this gene. |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows FBXO4 related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| BF331537 | NUP210 | 13 | 115 | 3 | 13392819 | 13392920 | FBXO4 | 107 | 317 | 5 | 41926160 | 41926370 | |
| BE073326 | NUP210 | 6 | 114 | 3 | 13392811 | 13392920 | FBXO4 | 106 | 316 | 5 | 41926160 | 41926370 | |
| BE073423 | NUP210 | 3 | 110 | 3 | 13392813 | 13392920 | FBXO4 | 102 | 312 | 5 | 41926160 | 41926370 | |
| Top |
| There's no copy number variation information in COSMIC data for this gene. |
| Top |
|
 |
| Top |
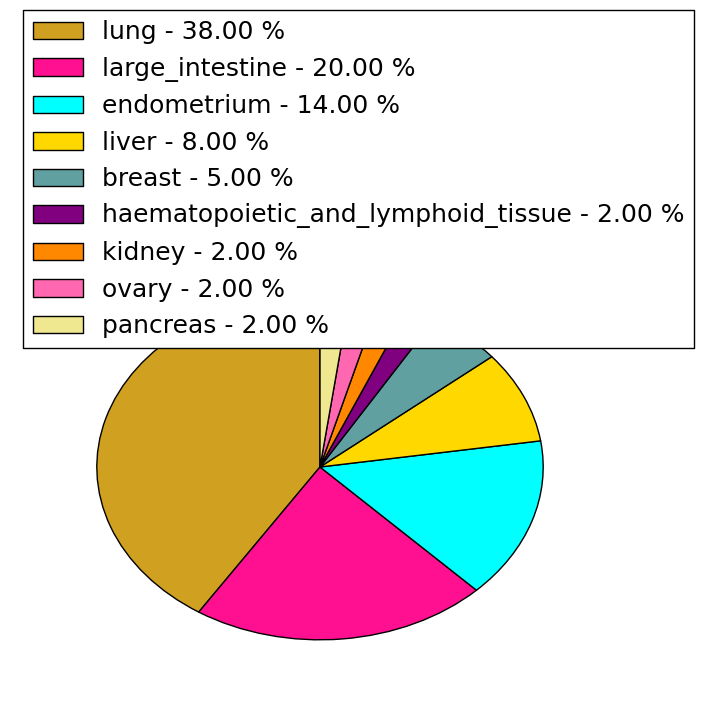
| Stat. for Non-Synonymous SNVs (# total SNVs=34) | (# total SNVs=9) |
 | 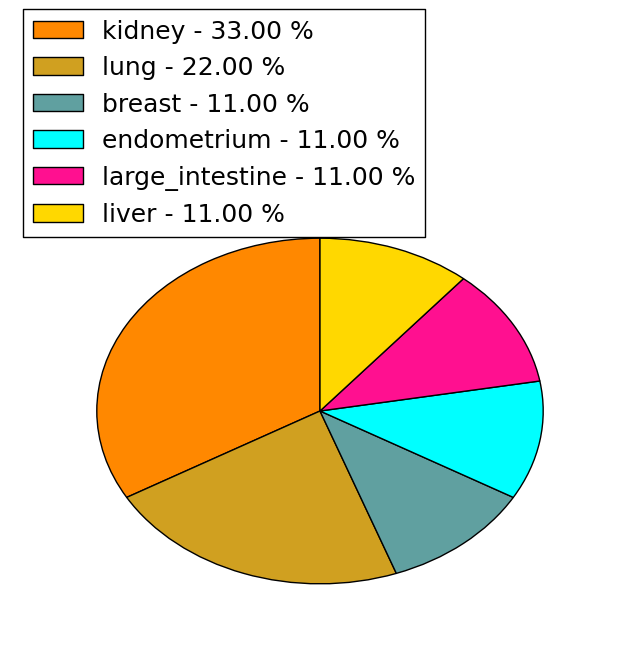 |
(# total SNVs=0) | (# total SNVs=0) |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr5:41941351-41941351 | p.L378I | 2 |
| chr5:41929935-41929935 | p.E188K | 2 |
| chr5:41941379-41941379 | p.R387K | 1 |
| chr5:41929976-41929976 | p.E201D | 1 |
| chr5:41934278-41934278 | p.N256H | 1 |
| chr5:41927273-41927273 | p.K116K | 1 |
| chr5:41939669-41939669 | p.F342S | 1 |
| chr5:41929858-41929858 | p.G162E | 1 |
| chr5:41941382-41941382 | p.*388L | 1 |
| chr5:41929978-41929978 | p.E202V | 1 |
| Top |
|
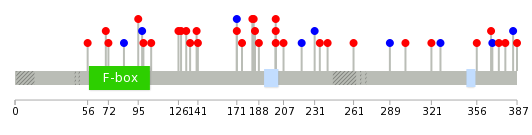 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 3 | 7 | 3 | 3 | 6 | 4 | 1 | 1 | 4 | 5 | ||||||||||
| # mutation | 3 | 7 | 3 | 3 | 9 | 4 | 1 | 1 | 5 | 5 | ||||||||||
| nonsynonymous SNV | 3 | 6 | 1 | 1 | 7 | 4 | 1 | 1 | 4 | 4 | ||||||||||
| synonymous SNV | 1 | 2 | 2 | 2 | 1 | 1 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr5:41929884 | p.L171L,FBXO4 | 2 |
| chr5:41934414 | p.R241I,FBXO4 | 1 |
| chr5:41927307 | p.A135V,FBXO4 | 1 |
| chr5:41929974 | p.R261L,FBXO4 | 1 |
| chr5:41939605 | p.M140L,FBXO4 | 1 |
| chr5:41927319 | p.F289F,FBXO4 | 1 |
| chr5:41929975 | p.A141T,FBXO4 | 1 |
| chr5:41925578 | p.K301R | 1 |
| chr5:41939628 | p.S321P | 1 |
| chr5:41927329 | p.N175S,FBXO4 | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
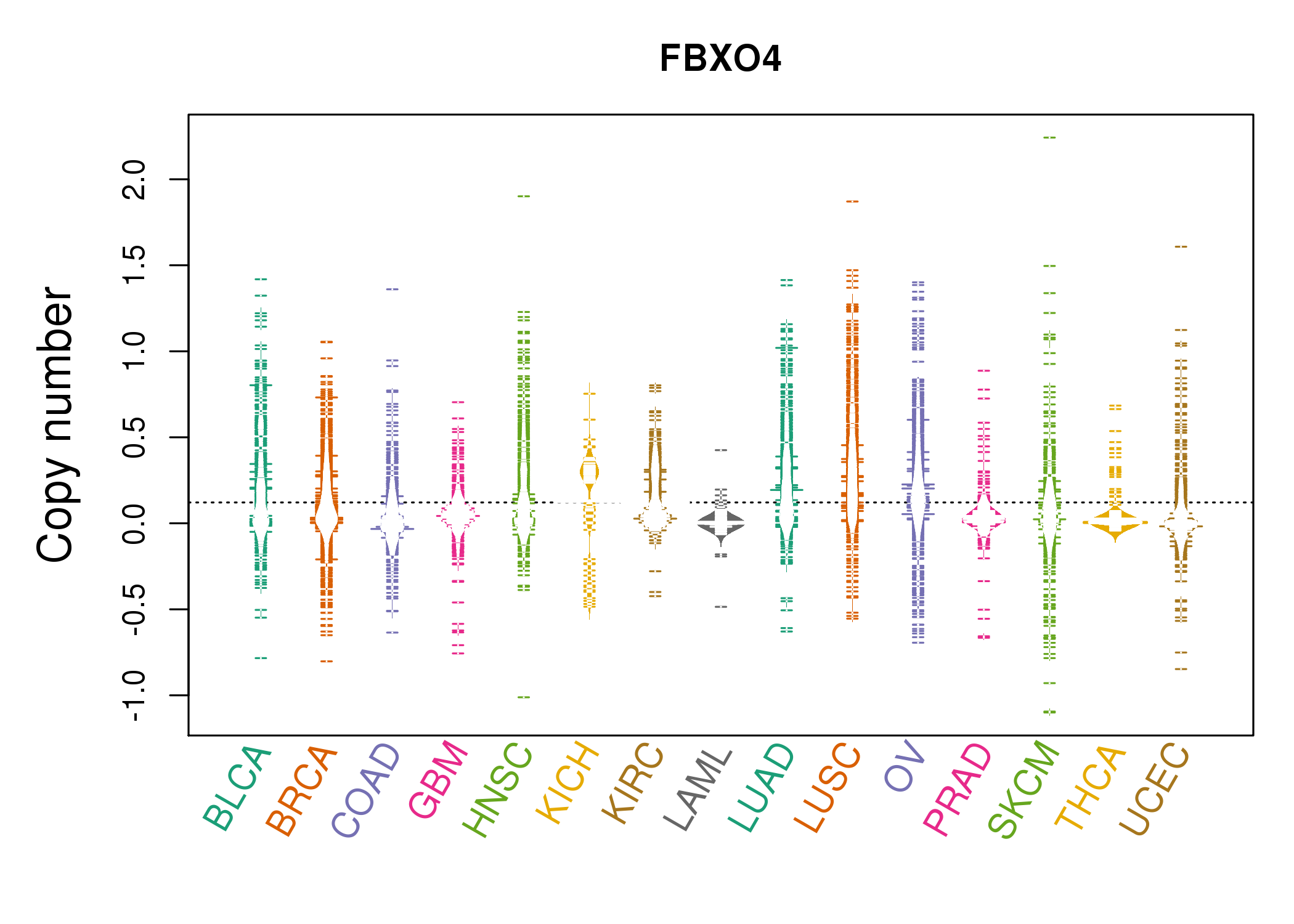 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
| Gene Expression for FBXO4 |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
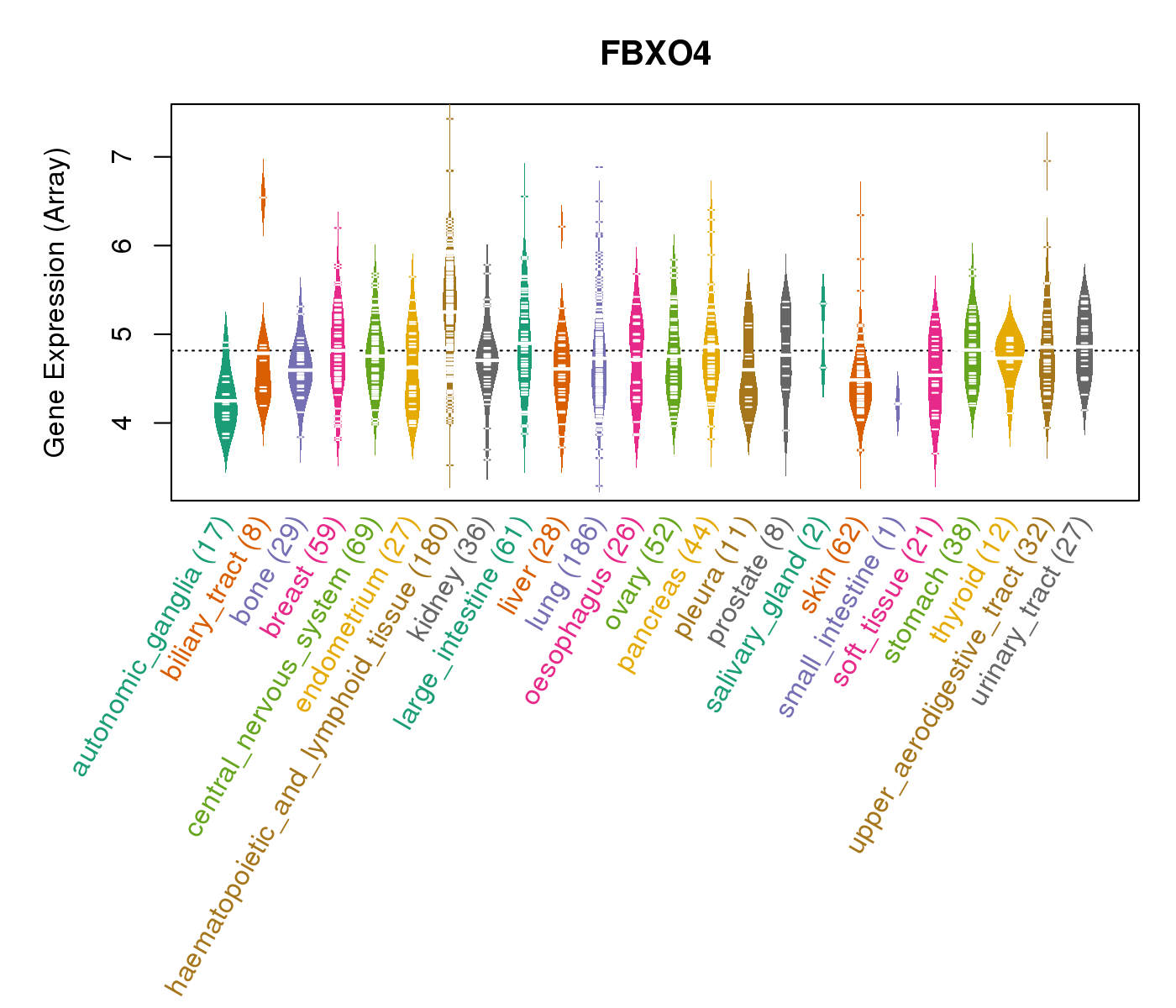 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
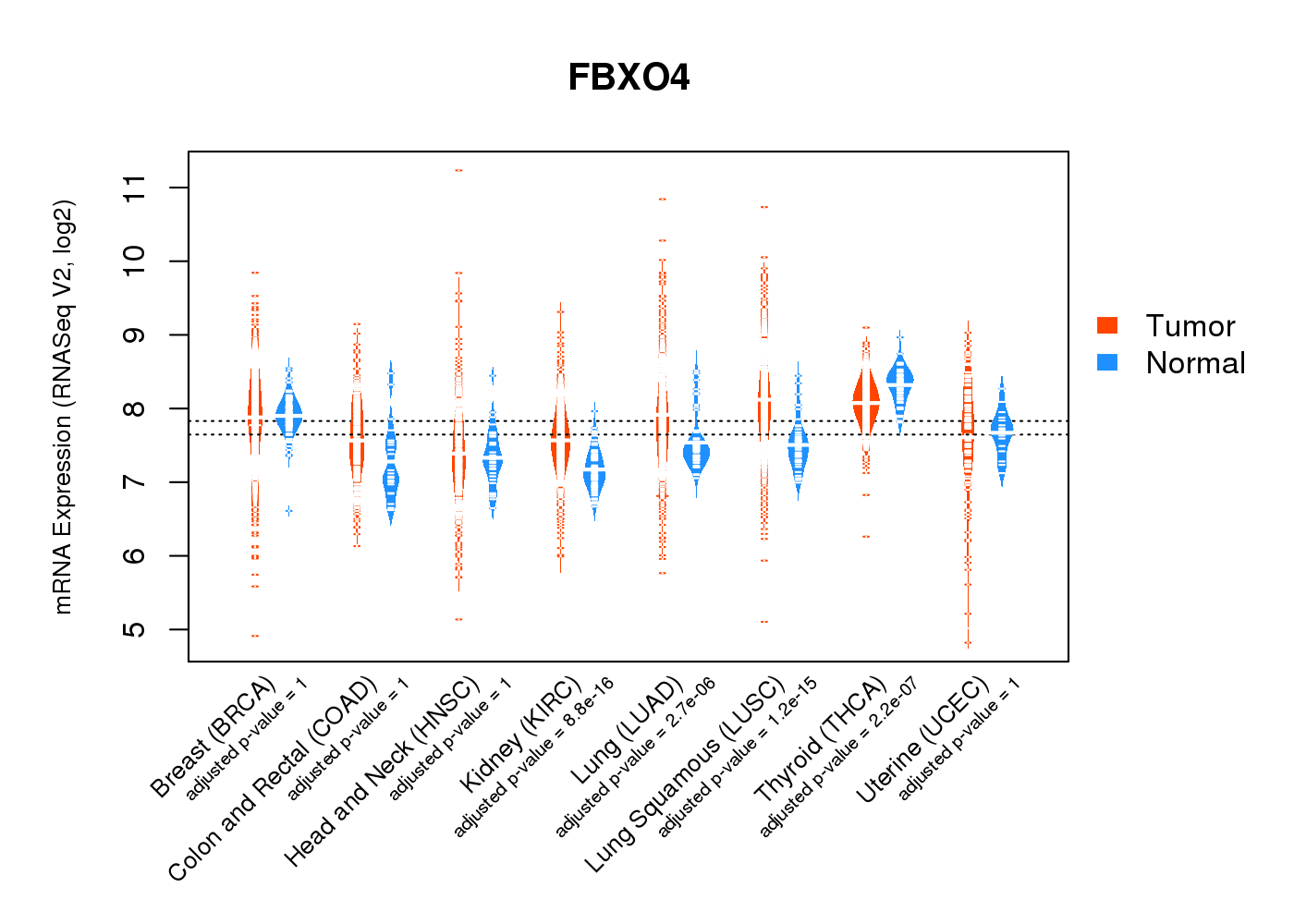 |
| Top |
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
 |
|
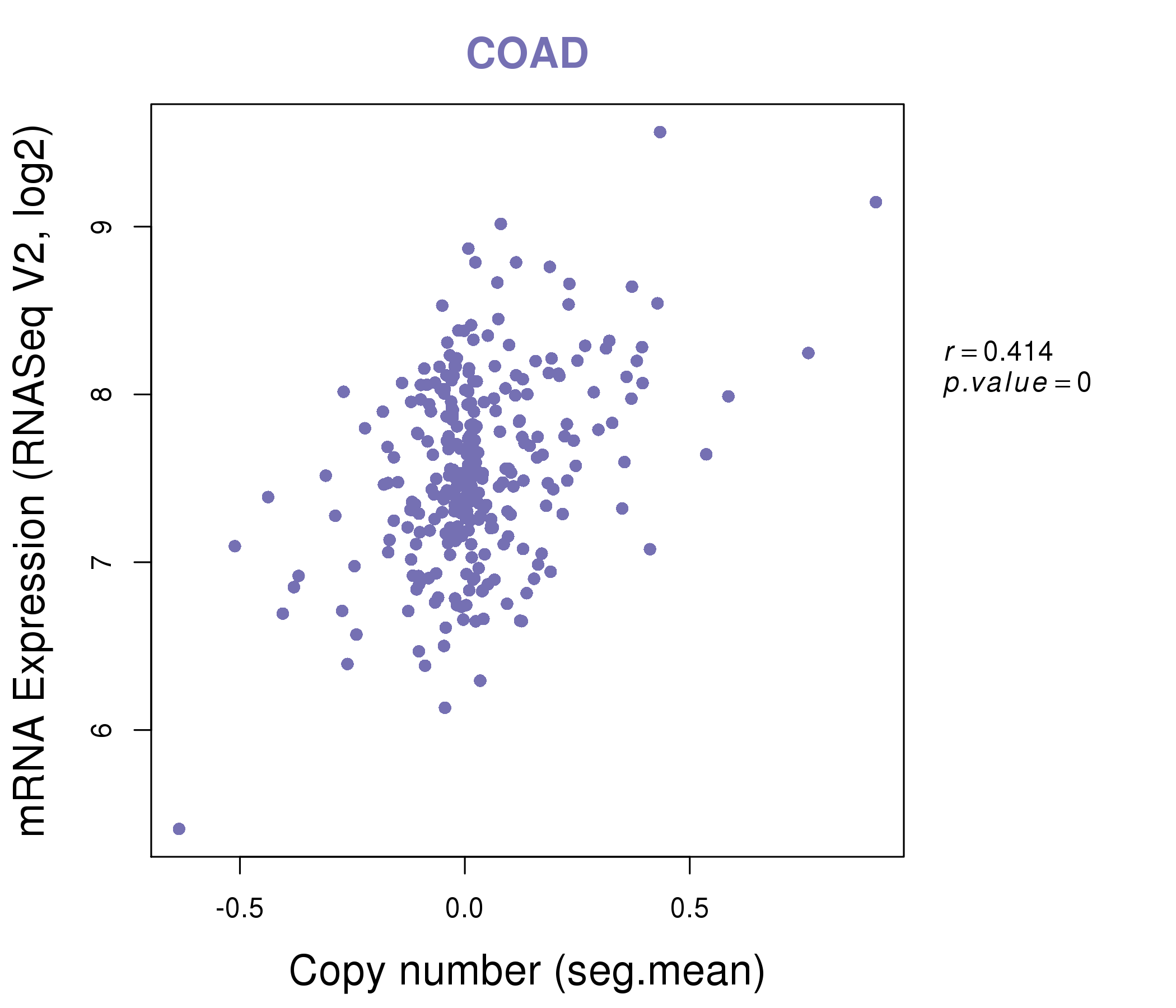 |
|
| Top |
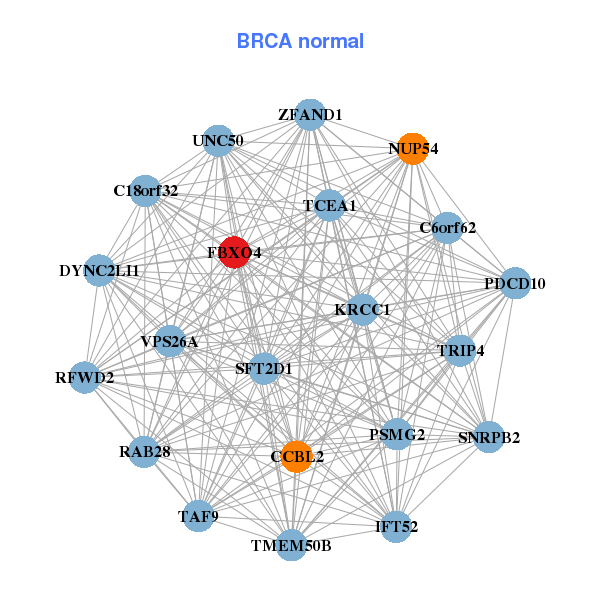
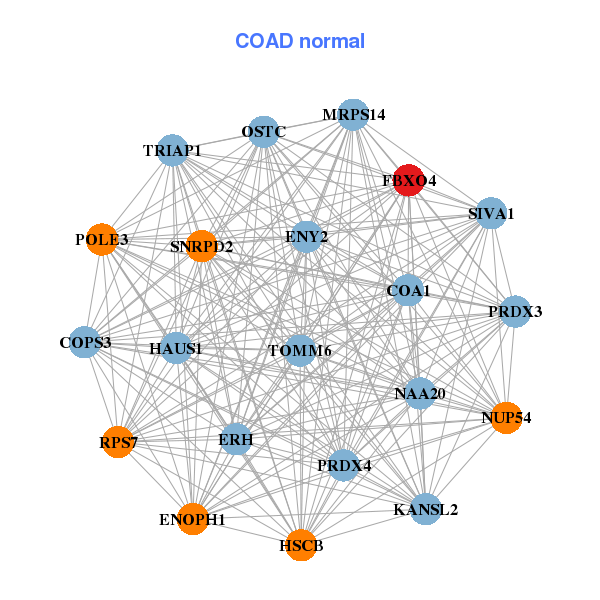
| Gene-Gene Network Information |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
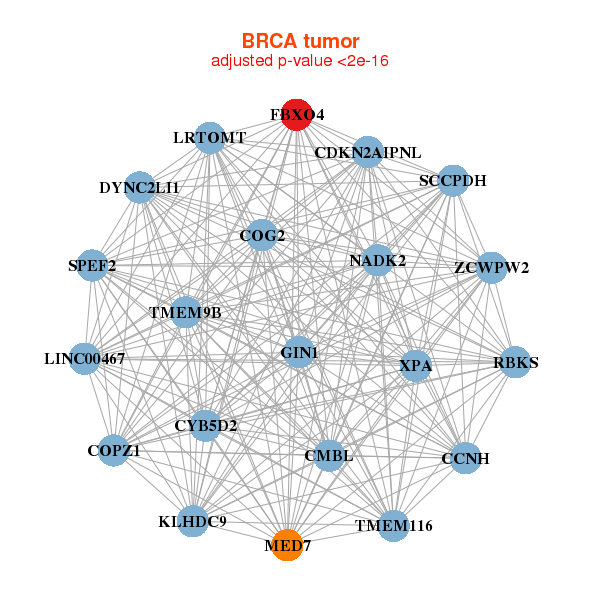 |
|
| LINC00467,NADK2,CCNH,CDKN2AIPNL,CMBL,COG2,COPZ1, CYB5D2,DYNC2LI1,FBXO4,GIN1,KLHDC9,LRTOMT,MED7, RBKS,SCCPDH,SPEF2,TMEM116,TMEM9B,XPA,ZCWPW2 | C18orf32,C6orf62,CCBL2,DYNC2LI1,FBXO4,IFT52,KRCC1, NUP54,PDCD10,PSMG2,RAB28,RFWD2,SFT2D1,SNRPB2, TAF9,TCEA1,TMEM50B,TRIP4,UNC50,VPS26A,ZFAND1 |
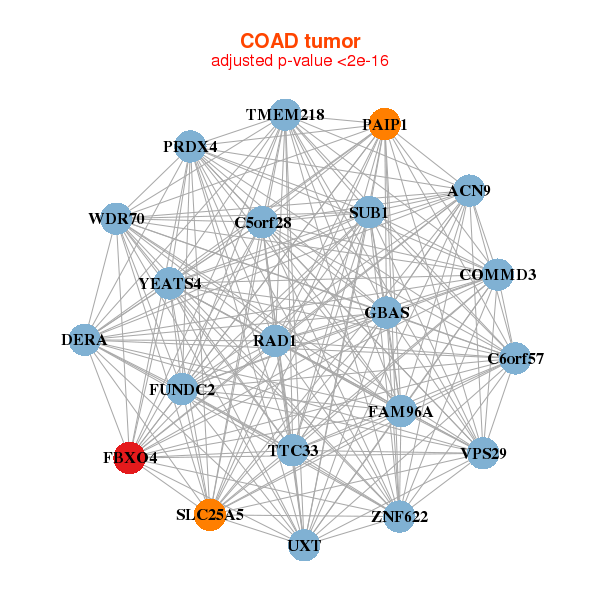 |
|
| ACN9,C5orf28,C6orf57,COMMD3,DERA,FAM96A,FBXO4, FUNDC2,GBAS,PAIP1,PRDX4,RAD1,SLC25A5,SUB1, TMEM218,TTC33,UXT,VPS29,WDR70,YEATS4,ZNF622 | KANSL2,COA1,COPS3,ENOPH1,ENY2,ERH,FBXO4, HAUS1,HSCB,MRPS14,NAA20,NUP54,OSTC,POLE3, PRDX3,PRDX4,RPS7,SIVA1,SNRPD2,TOMM6,TRIAP1 |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for FBXO4 |
| There's no related Drug. |
| Top |
| Cross referenced IDs for FBXO4 |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |