|
||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for ACHE |
| Top |
| Phenotypic Information for ACHE(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: ACHE |
| Familial Cancer Database: ACHE | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
| . | ||||||||||||||
Oncogene 1 | Significant driver gene in | |||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| KEGG_GLYCEROPHOSPHOLIPID_METABOLISM REACTOME_PHOSPHOLIPID_METABOLISM REACTOME_METABOLISM_OF_LIPIDS_AND_LIPOPROTEINS | |
| OMIM | |
| Orphanet | |
| Disease | KEGG Disease: ACHE |
| MedGen: ACHE (Human Medical Genetics with Condition) | |
| ClinVar: ACHE | |
| Phenotype | MGI: ACHE (International Mouse Phenotyping Consortium) |
| PhenomicDB: ACHE | |
| Mutations for ACHE |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
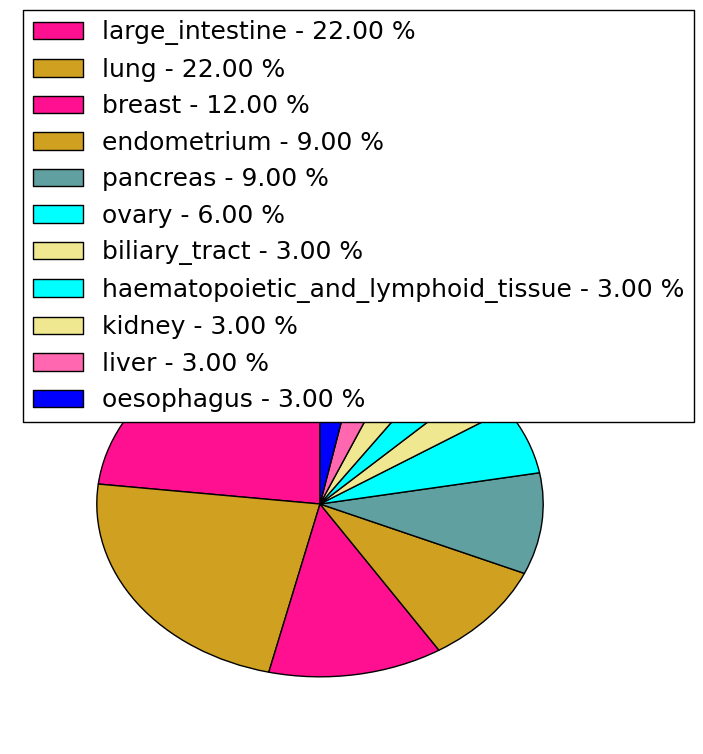
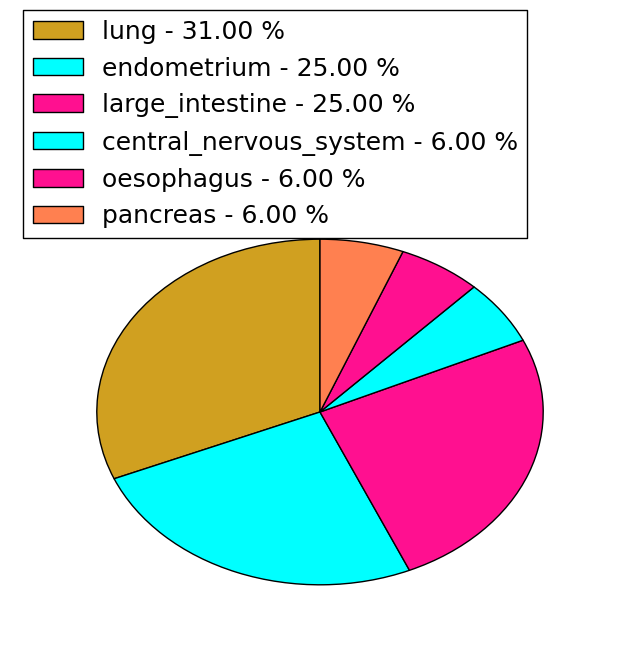
| - Statistics for Tissue and Mutation type | Top |
 |
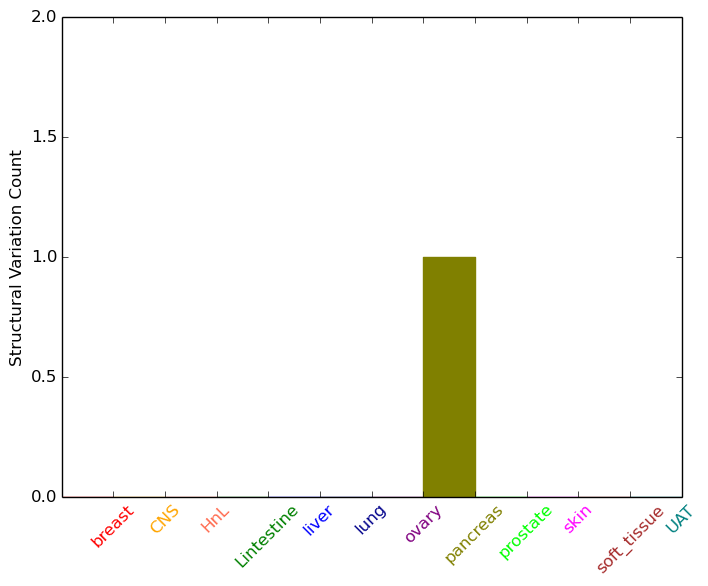
| - For Inter-chromosomal Variations |
| * Inter-chromosomal variantions includes 'interchromosomal amplicon to amplicon', 'interchromosomal amplicon to non-amplified dna', 'interchromosomal insertion', 'Interchromosomal unknown type'. |
 |
| - For Intra-chromosomal Variations |
| There's no intra-chromosomal structural variation. |
| Sample | Symbol_a | Chr_a | Start_a | End_a | Symbol_b | Chr_b | Start_b | End_b |
| pancreas | ACHE | chr7 | 100489340 | 100489360 | chr15 | 83421902 | 83421922 |
| cf) Tissue number; Tissue name (1;Breast, 2;Central_nervous_system, 3;Haematopoietic_and_lymphoid_tissue, 4;Large_intestine, 5;Liver, 6;Lung, 7;Ovary, 8;Pancreas, 9;Prostate, 10;Skin, 11;Soft_tissue, 12;Upper_aerodigestive_tract) |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows ACHE related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| R96128 | CDK6 | 1 | 44 | 7 | 92331061 | 92331104 | ACHE | 45 | 382 | 7 | 100490125 | 100490803 | |
| Top |
| There's no copy number variation information in COSMIC data for this gene. |
| Top |
|
 |
| Top |
| Stat. for Non-Synonymous SNVs (# total SNVs=31) | (# total SNVs=16) |
 |  |
(# total SNVs=0) | (# total SNVs=0) |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr7:100491611-100491611 | p.P81P | 3 |
| chr7:100490067-100490067 | p.E481K | 3 |
| chr7:100491150-100491150 | p.A235E | 2 |
| chr7:100488658-100488658 | p.P592R | 2 |
| chr7:100491461-100491461 | p.N131N | 2 |
| chr7:100491616-100491616 | p.P80S | 2 |
| chr7:100490875-100490875 | p.R327W | 2 |
| chr7:100490077-100490077 | p.P477P | 2 |
| chr7:100489954-100489954 | p.? | 1 |
| chr7:100490900-100490900 | p.H318H | 1 |
| Top |
|
 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 5 | 1 | 7 | 1 | 3 | 7 | 4 | 1 | 5 | 13 | 7 | |||||||||
| # mutation | 6 | 1 | 7 | 1 | 3 | 7 | 4 | 1 | 5 | 14 | 7 | |||||||||
| nonsynonymous SNV | 4 | 4 | 2 | 4 | 3 | 1 | 4 | 11 | 3 | |||||||||||
| synonymous SNV | 2 | 1 | 3 | 1 | 1 | 3 | 1 | 1 | 3 | 4 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr7:100491048 | p.E481K,ACHE | 2 |
| chr7:100491461 | p.T269M,ACHE | 2 |
| chr7:100490067 | p.P80S,ACHE | 2 |
| chr7:100491616 | p.N131N,ACHE | 2 |
| chr7:100490017 | p.I482V,ACHE | 1 |
| chr7:100491327 | p.H318H,ACHE | 1 |
| chr7:100490391 | p.E125E,ACHE | 1 |
| chr7:100491627 | p.H315H,ACHE | 1 |
| chr7:100487859 | p.D105Y,ACHE | 1 |
| chr7:100490047 | p.T467M,ACHE | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
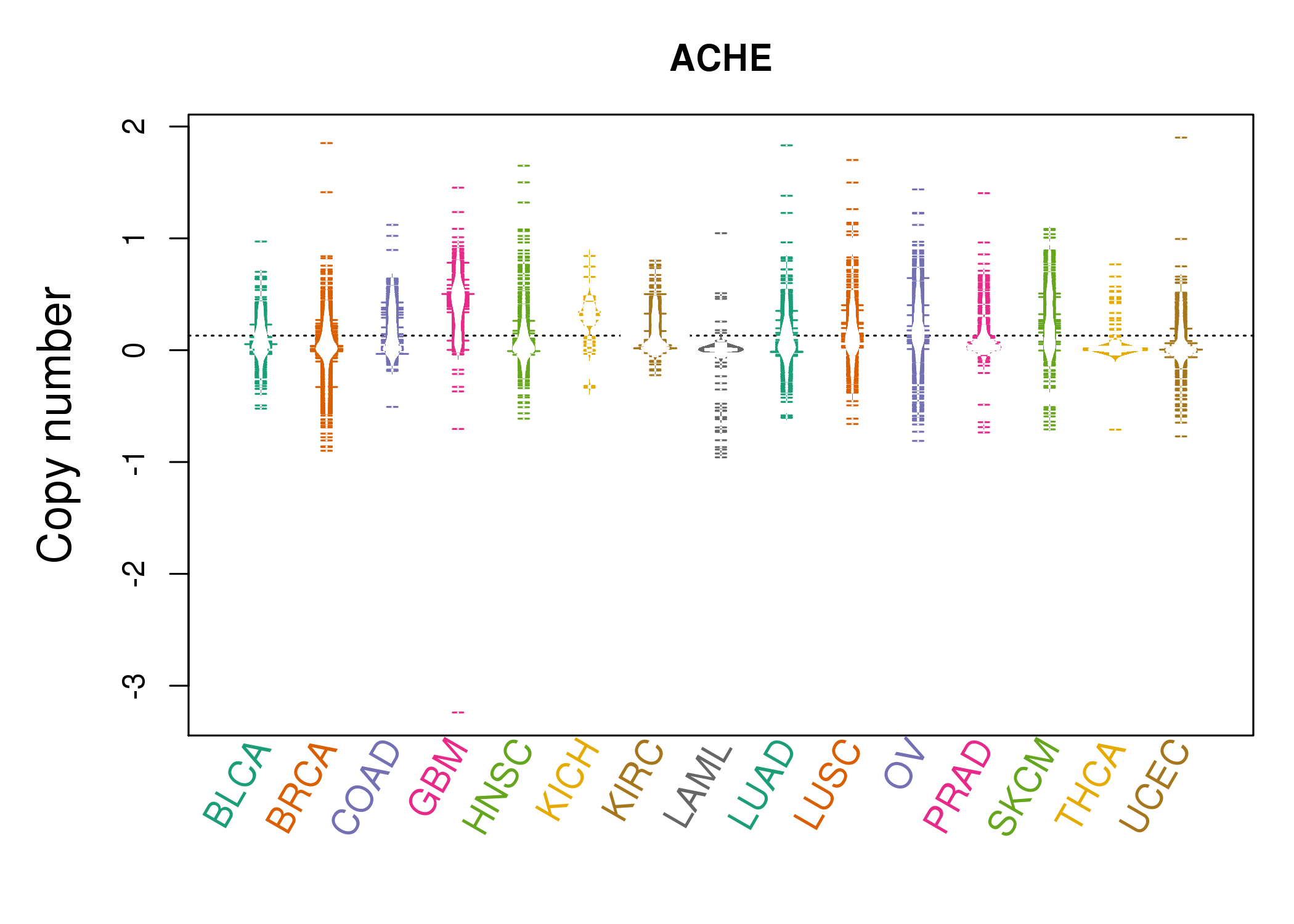 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
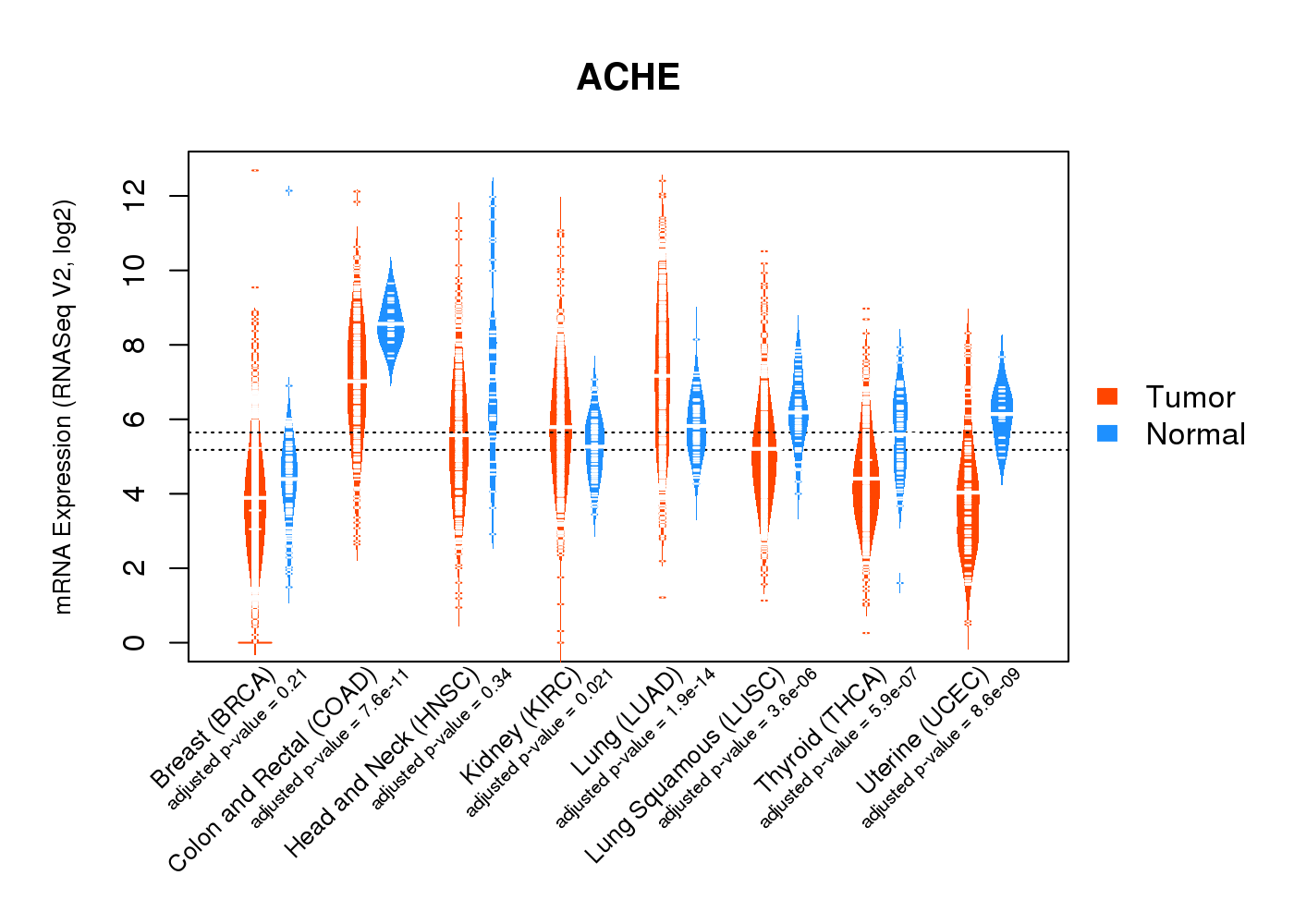
| Gene Expression for ACHE |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
 |
| Top |
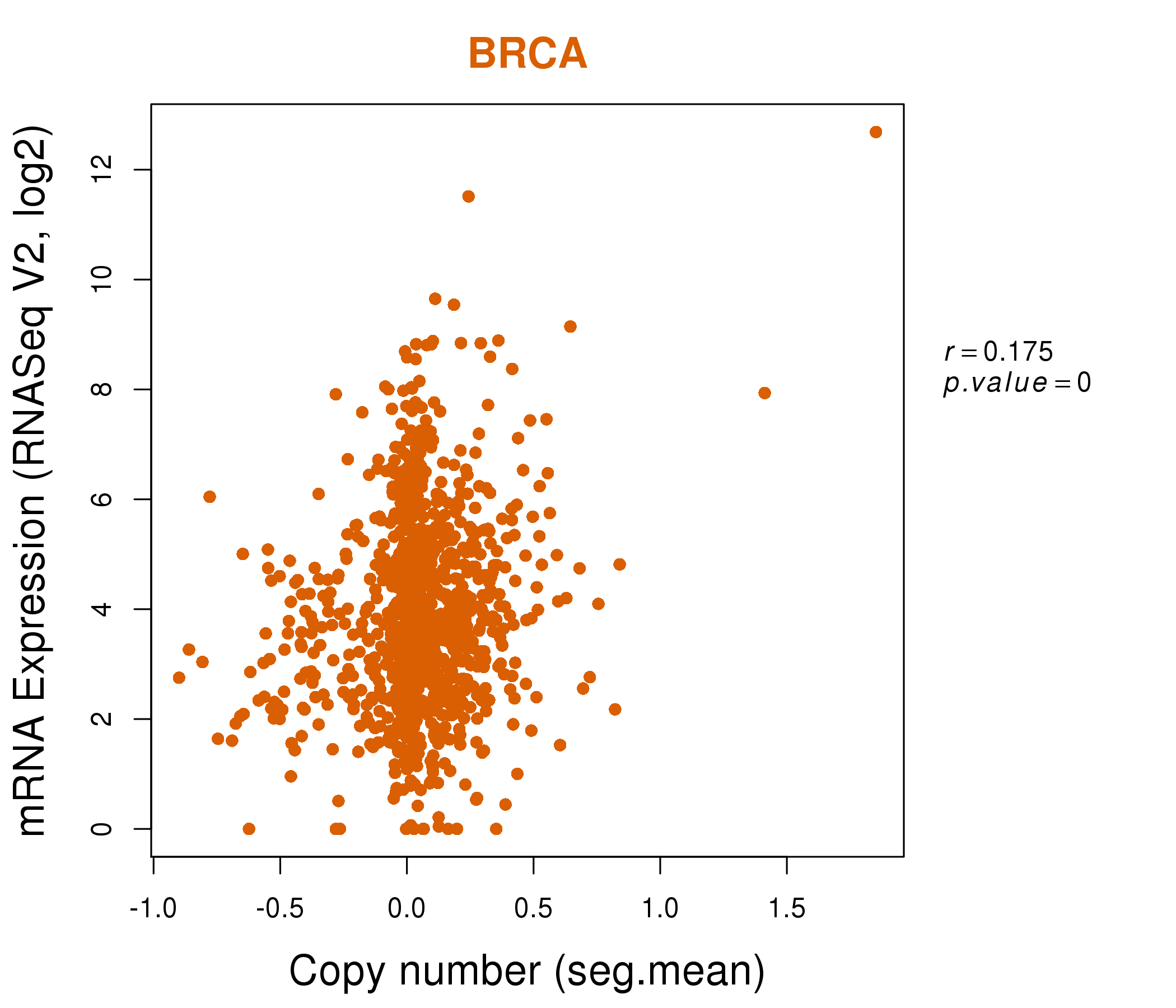
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
 |
|
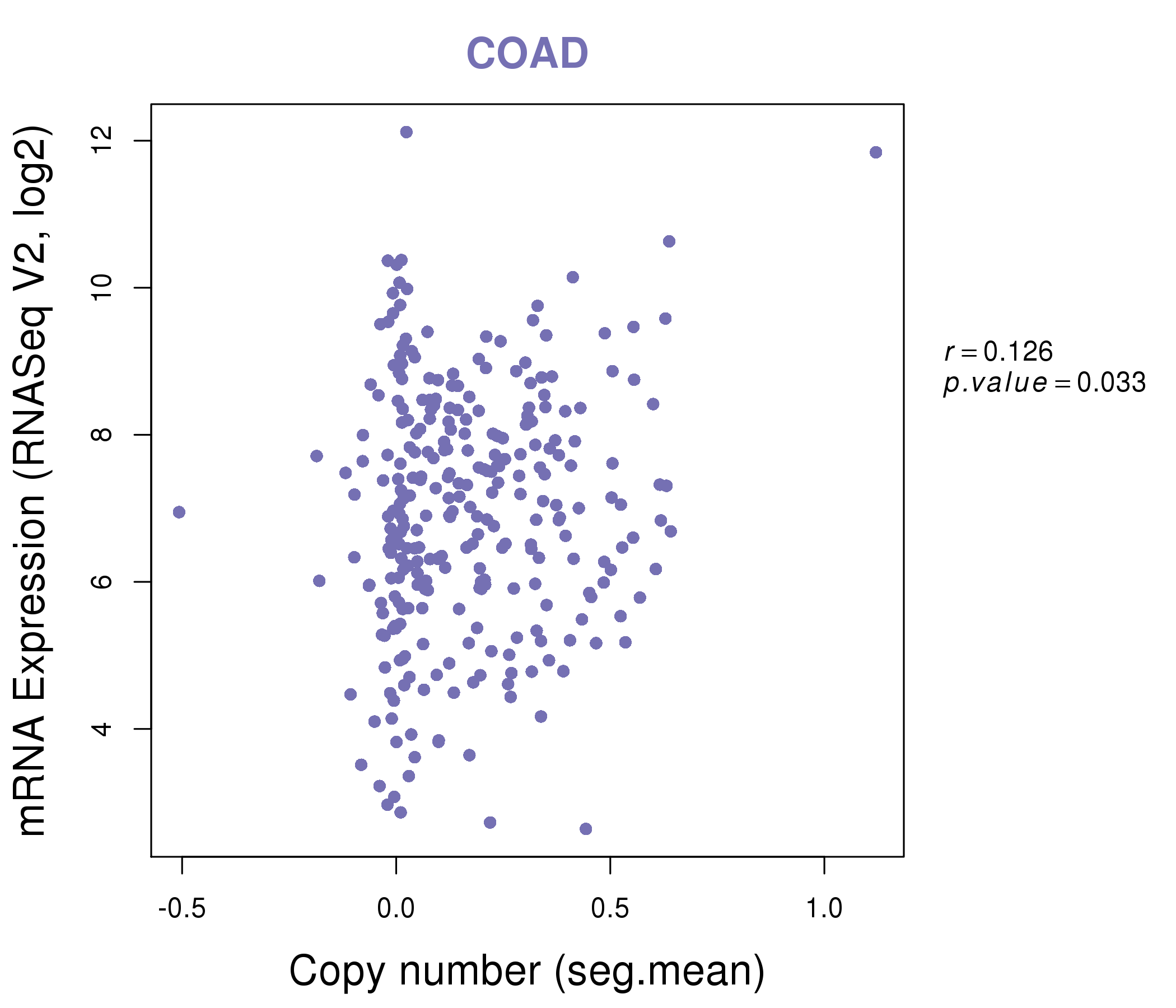 |
|
| Top |
| Gene-Gene Network Information |
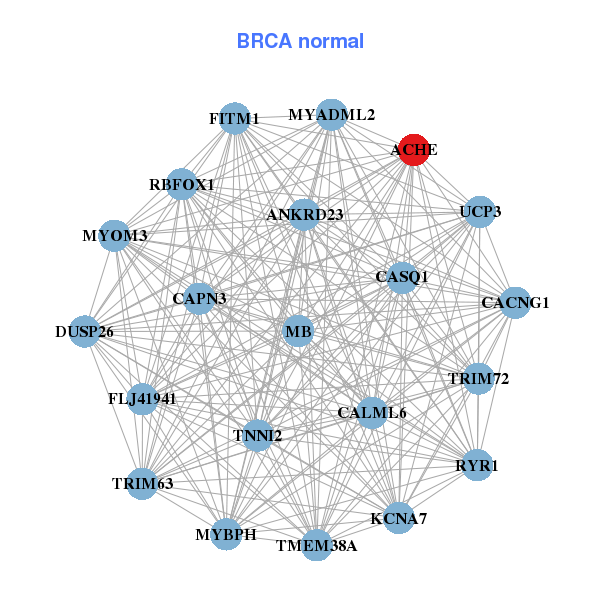
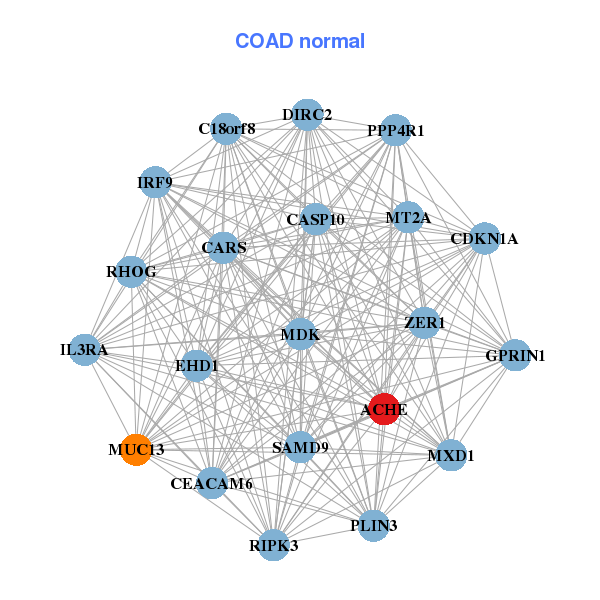
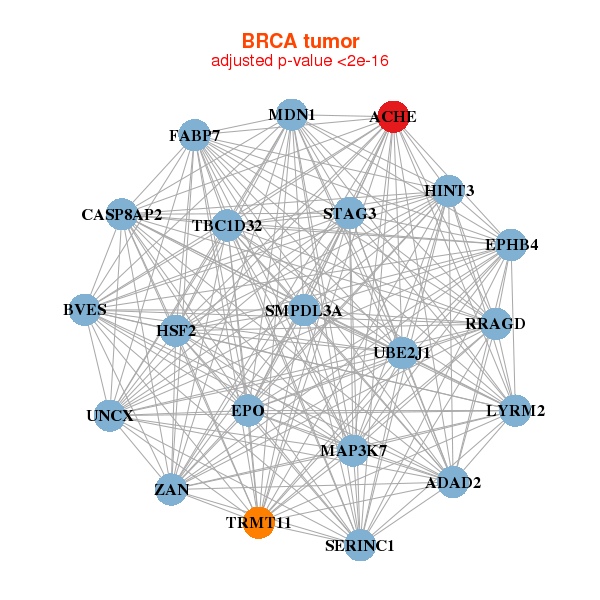
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
 |
| ||||
| ACHE,ADAD2,BVES,TBC1D32,CASP8AP2,EPHB4,EPO, FABP7,HINT3,HSF2,LYRM2,MAP3K7,MDN1,RRAGD, SERINC1,SMPDL3A,STAG3,TRMT11,UBE2J1,UNCX,ZAN | RBFOX1,ACHE,ANKRD23,CACNG1,CALML6,CAPN3,CASQ1, DUSP26,FITM1,FLJ41941,KCNA7,MB,MYADML2,MYBPH, MYOM3,RYR1,TMEM38A,TNNI2,TRIM63,TRIM72,UCP3 | ||||
 |
| ||||
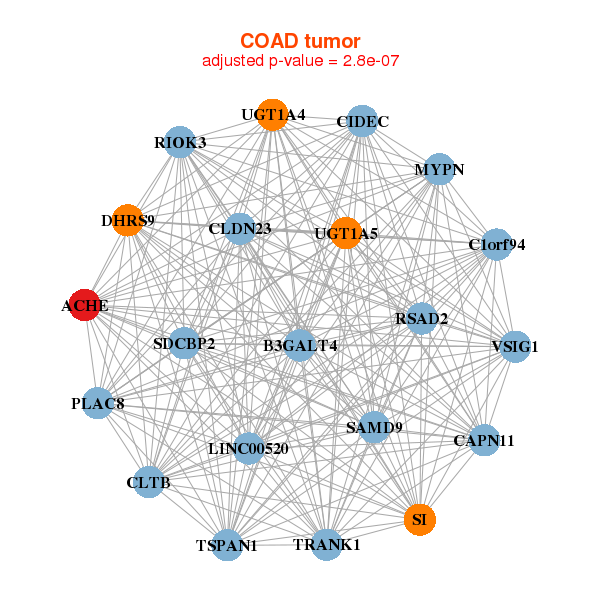
| ACHE,B3GALT4,LINC00520,C1orf94,CAPN11,CIDEC,CLDN23, CLTB,DHRS9,MYPN,PLAC8,RIOK3,RSAD2,SAMD9, SDCBP2,SI,TRANK1,TSPAN1,UGT1A4,UGT1A5,VSIG1 | ACHE,C18orf8,CARS,CASP10,CDKN1A,CEACAM6,DIRC2, EHD1,GPRIN1,IL3RA,IRF9,MDK,MT2A,MUC13, MXD1,PLIN3,PPP4R1,RHOG,RIPK3,SAMD9,ZER1 |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for ACHE |
| DB Category | DB Name | DB's ID and Url link |
| * Gene Centered Interaction Network. |
 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB00122 | acetylcholinesterase | approved; nutraceutical | Choline | 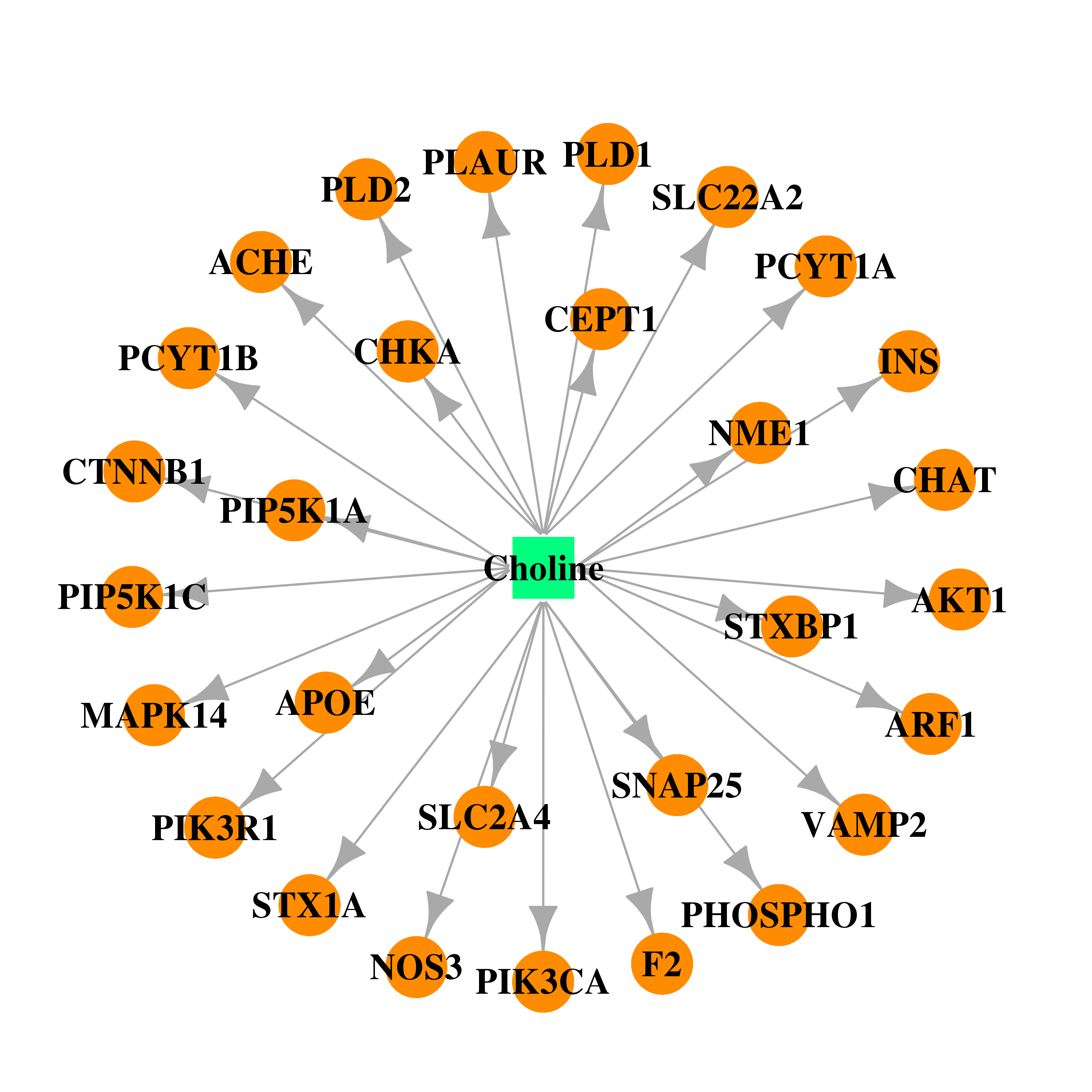 | 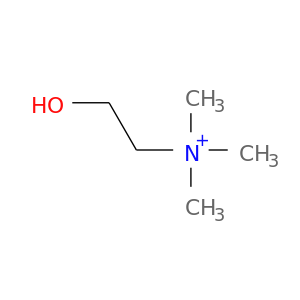 |
| DB00382 | acetylcholinesterase | approved | Tacrine |  | 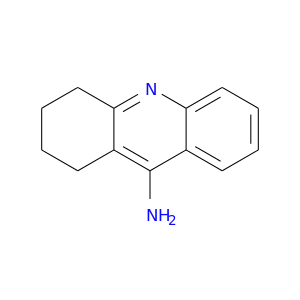 |
| DB00483 | acetylcholinesterase | approved | Gallamine Triethiodide | 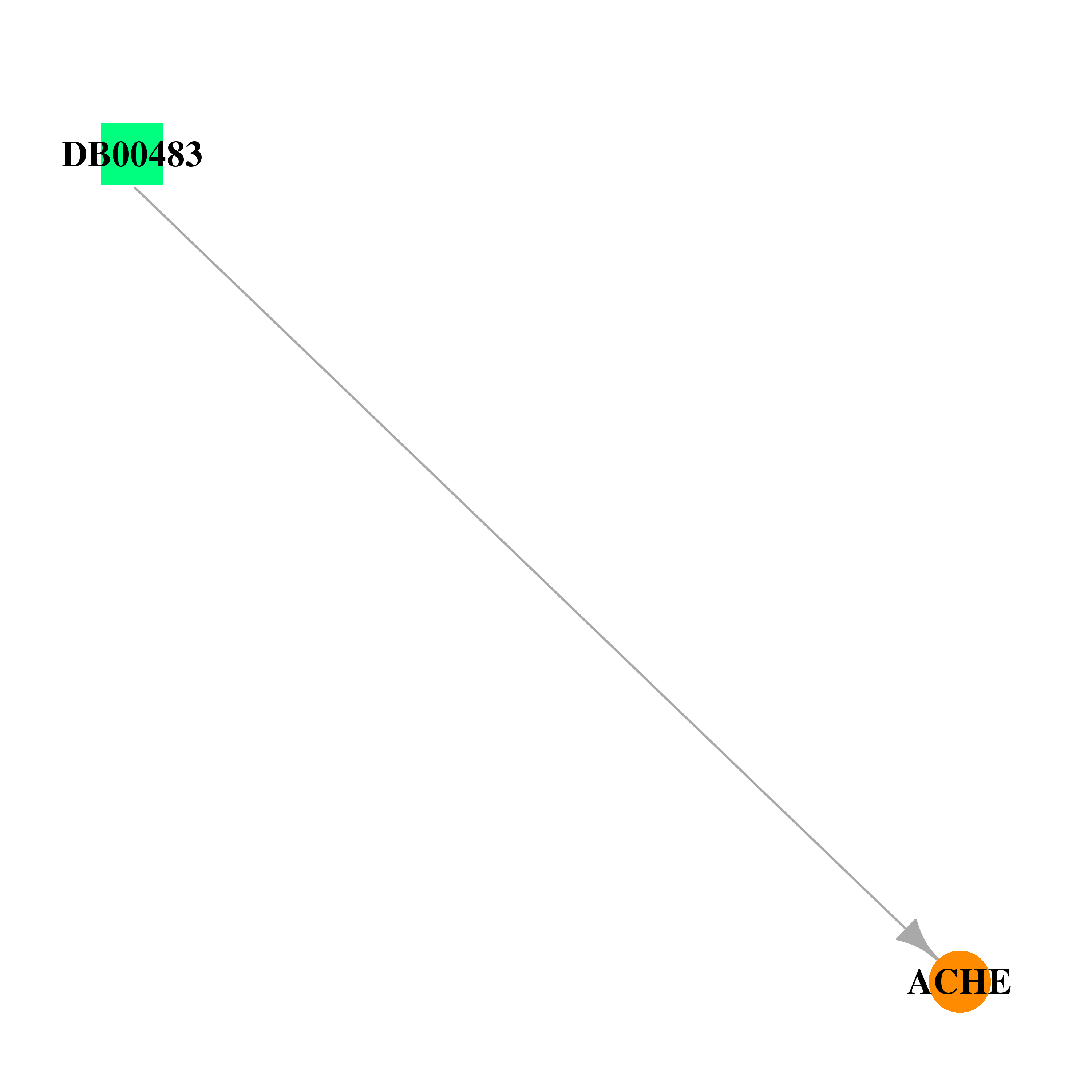 |  |
| DB00545 | acetylcholinesterase | approved | Pyridostigmine | 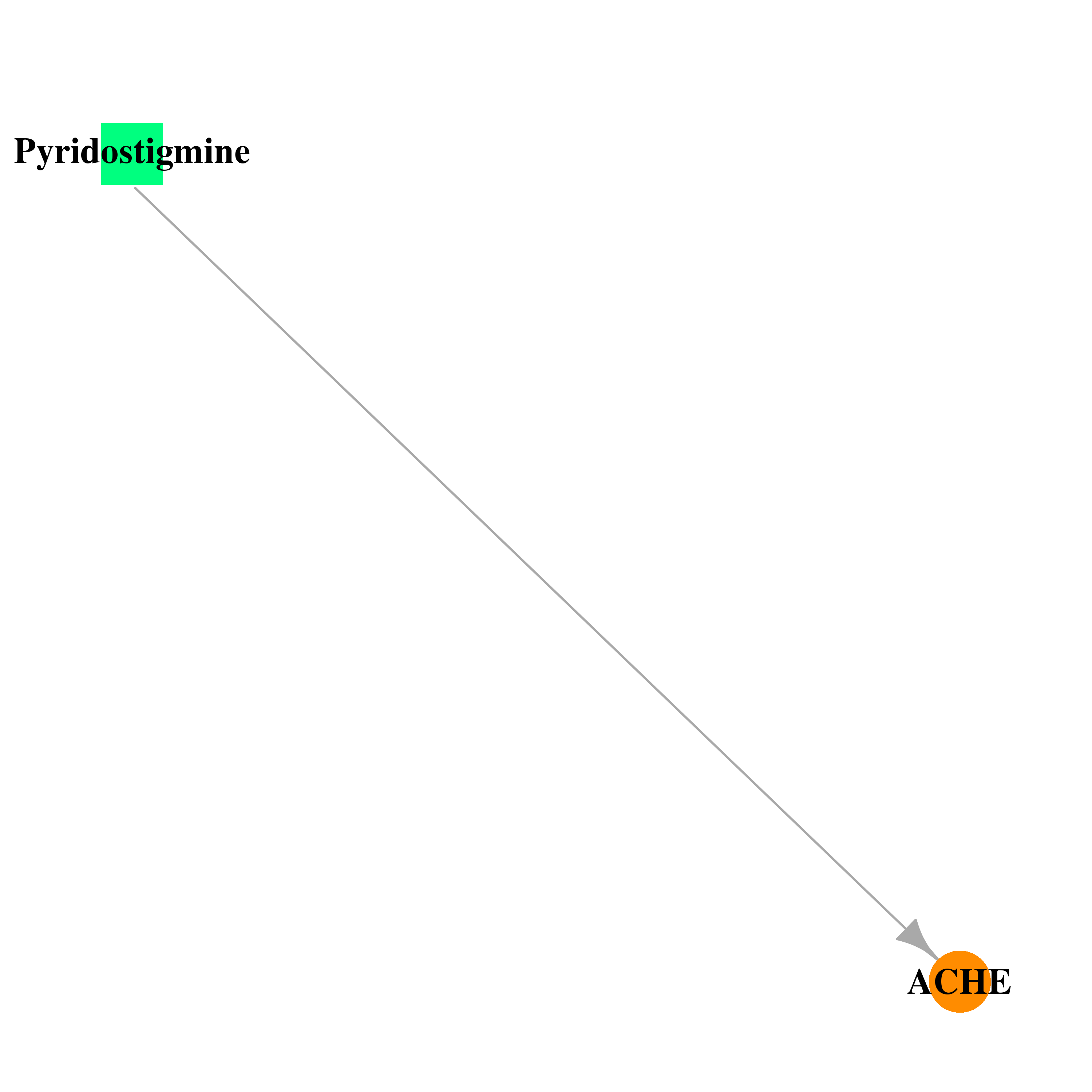 |  |
| DB00674 | acetylcholinesterase | approved | Galantamine | 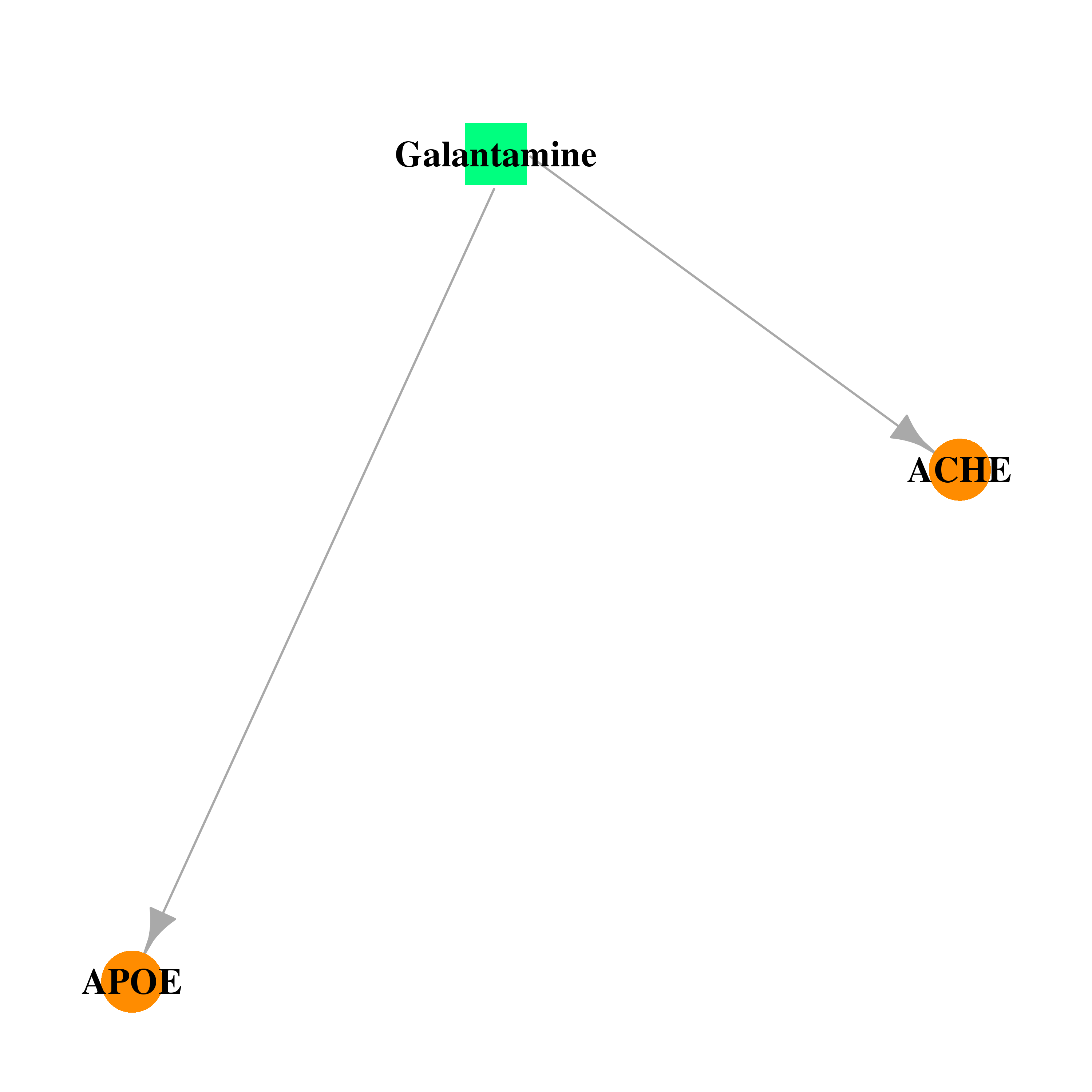 |  |
| DB00677 | acetylcholinesterase | approved | Isoflurophate |  |  |
| DB00733 | acetylcholinesterase | approved | Pralidoxime |  |  |
| DB00805 | acetylcholinesterase | approved | Minaprine | 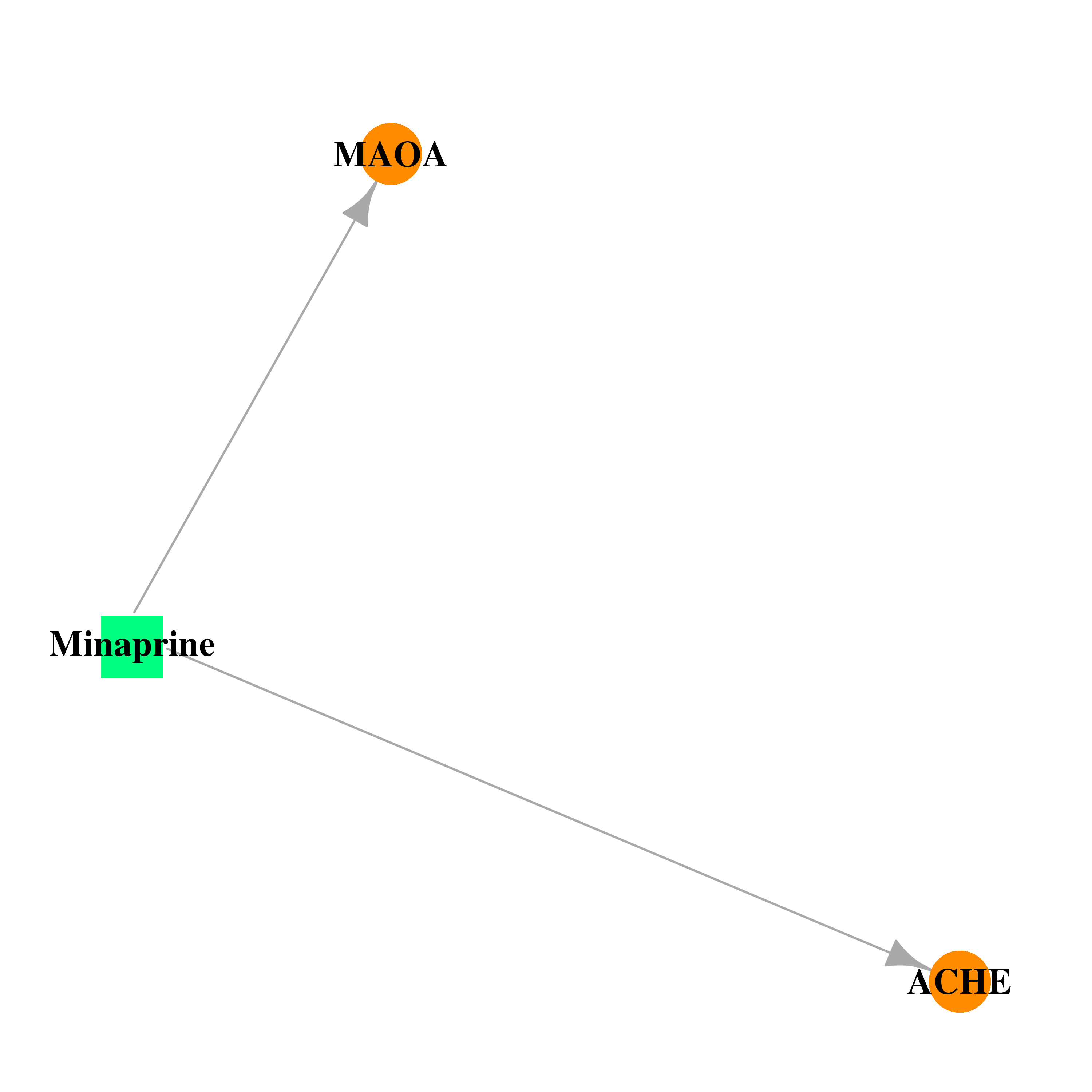 |  |
| DB00843 | acetylcholinesterase | approved | Donepezil | 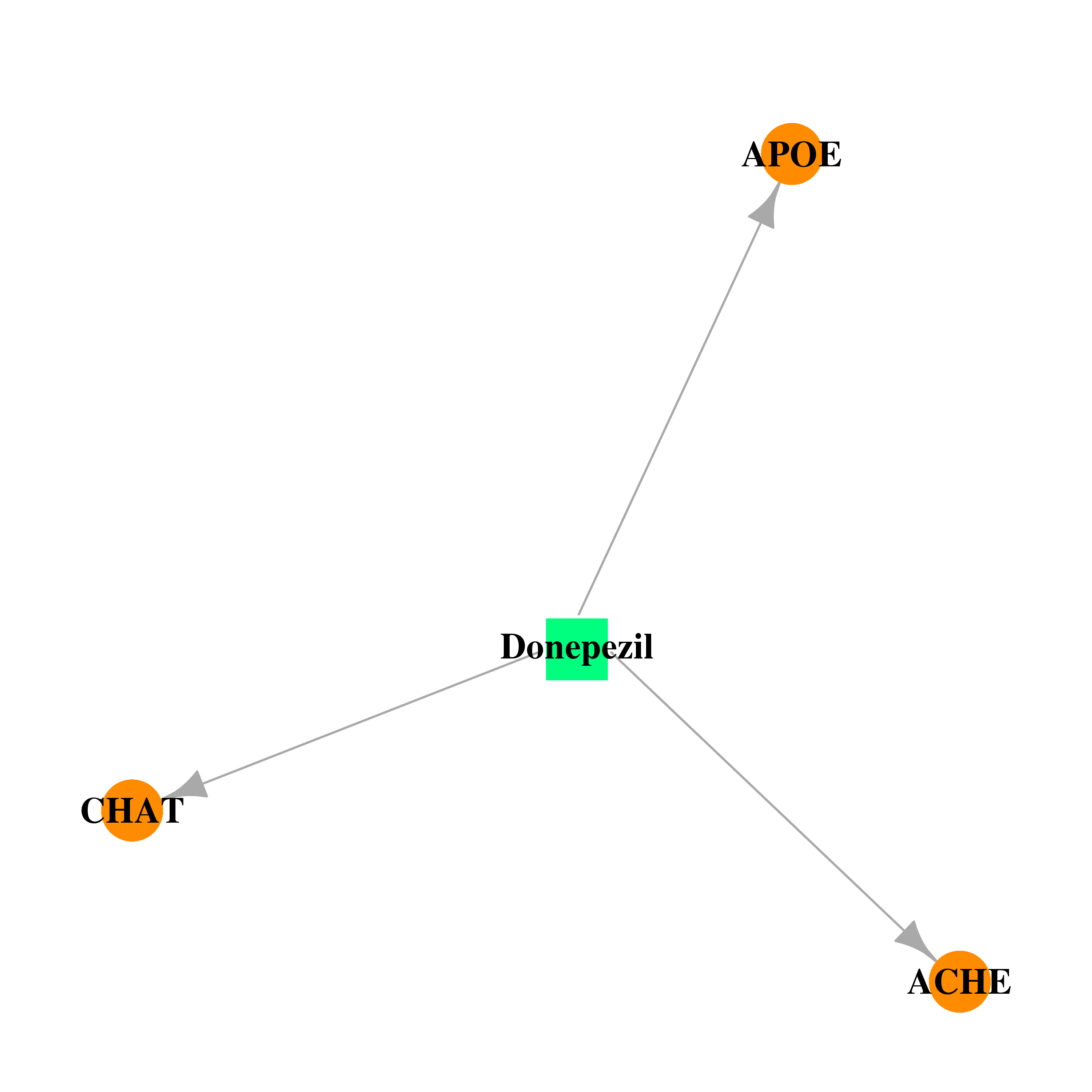 |  |
| DB00944 | acetylcholinesterase | approved | Demecarium |  |  |
| DB00981 | acetylcholinesterase | approved | Physostigmine |  |  |
| DB00989 | acetylcholinesterase | approved; investigational | Rivastigmine |  |  |
| DB01010 | acetylcholinesterase | approved | Edrophonium |  |  |
| DB01122 | acetylcholinesterase | approved | Ambenonium |  |  |
| DB01199 | acetylcholinesterase | approved | Tubocurarine |  |  |
| DB01245 | acetylcholinesterase | approved | Decamethonium |  |  |
| DB01364 | acetylcholinesterase | approved | Ephedrine |  |  |
| DB01400 | acetylcholinesterase | approved | Neostigmine | 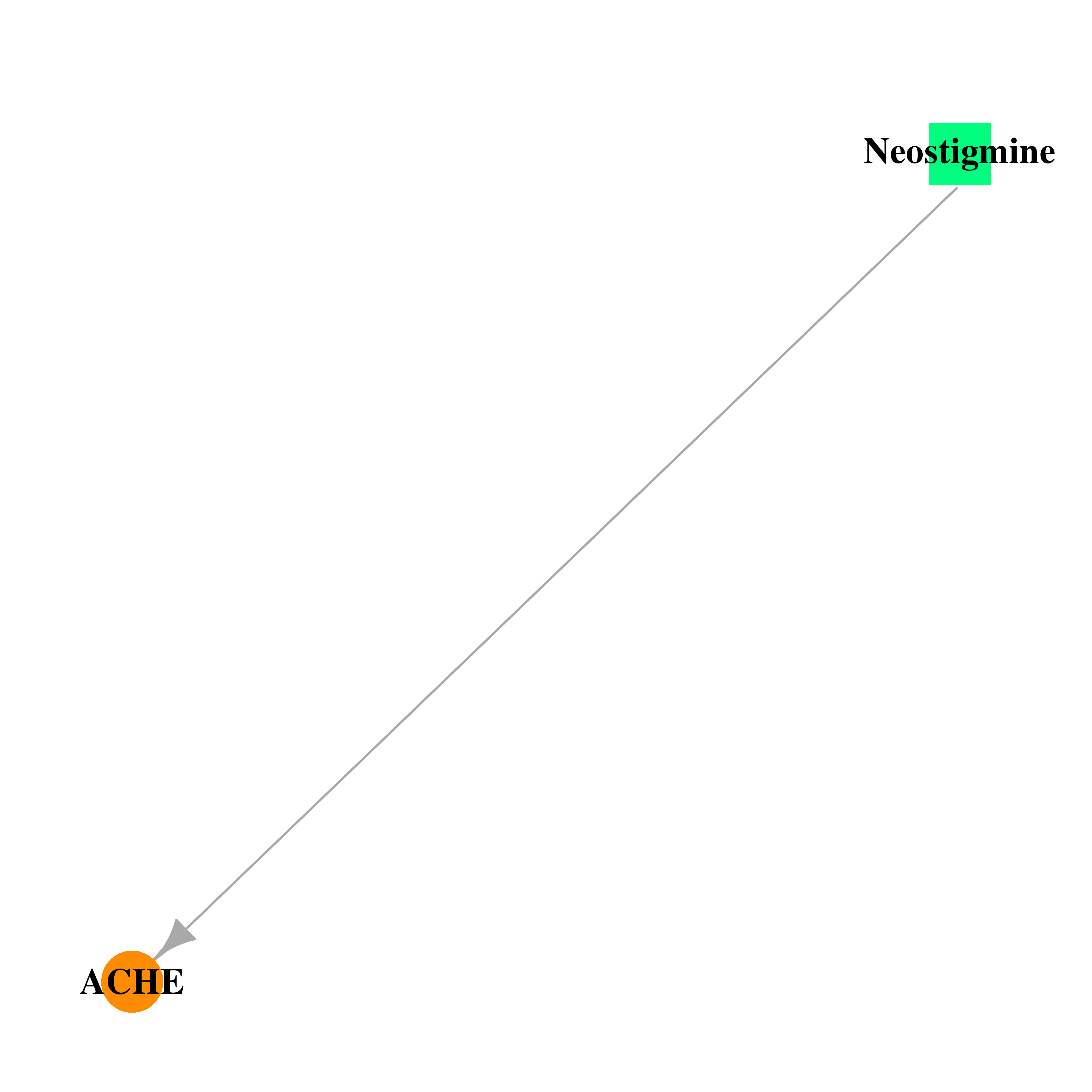 |  |
| DB01805 | acetylcholinesterase | experimental | Monoisopropylphosphorylserine |  |  |
| DB01928 | acetylcholinesterase | experimental | Huperaine A |  | 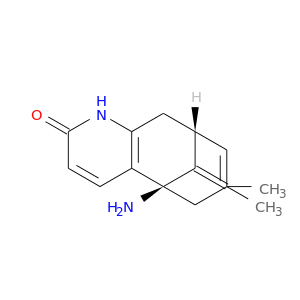 |
| DB02166 | acetylcholinesterase | experimental | Propidium |  | 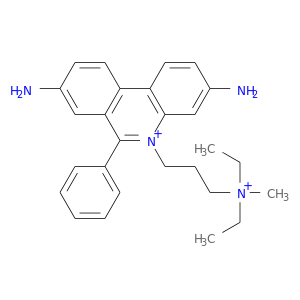 |
| DB02226 | acetylcholinesterase | experimental | 3,8-Diamino-6-Phenyl-5-[6-[1-[2-[(1,2,3,4-Tetrahydro-9-Acridinyl)Amino]Ethyl]-1h-1,2,3-Triazol-4-Yl]Hexyl]-Phenanthridinium |  |  |
| DB02343 | acetylcholinesterase | experimental | 3,6,9,12,15-Pentaoxaheptadecane |  |  |
| DB02404 | acetylcholinesterase | experimental | 1-Deoxy-1-Thio-Heptaethylene Glycol |  |  |
| DB02673 | acetylcholinesterase | experimental | (4ar,6s,8ar)-11-[8-(1,3-Dioxo-1,3-Dihydro-2h-Isoindol-2-Yl)Octyl]-6-Hydroxy-3-Methoxy-5,6,9,10-Tetrahydro-4ah-[1]Benzofuro[3a,3,2-Ef][2]Benzazepin-11-Ium |  |  |
| DB02825 | acetylcholinesterase | experimental | Methylphosphonic Acid Ester Group |  |  |
| DB02845 | acetylcholinesterase | experimental | Methylphosphinic Acid |  |  |
| DB03005 | acetylcholinesterase | experimental | 3,8-Diamino-6-Phenyl-5-[6-[1-[2-[(1,2,3,4-Tetrahydro-9-Acridinyl)Amino]Ethyl]-1h-1,2,3-Triazol-5-Yl]Hexyl]-Phenanthridinium |  |  |
| DB03128 | acetylcholinesterase | experimental | Acetylcholine |  |  |
| DB03283 | acetylcholinesterase | experimental | beta-L-fucose |  |  |
| DB03359 | acetylcholinesterase | experimental | M-(N,N,N-Trimethylammonio)-2,2,2-Trifluoro-1,1-Dihydroxyethylbenzene |  |  |
| DB03740 | acetylcholinesterase | experimental | 2-(Acetylamino)-2-Deoxy-a-D-Glucopyranose |  |  |
| DB03814 | acetylcholinesterase | experimental | 2-(N-Morpholino)-Ethanesulfonic Acid |  | 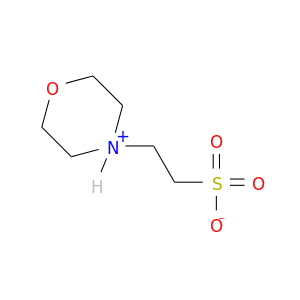 |
| DB04021 | acetylcholinesterase | experimental | MF268 |  | 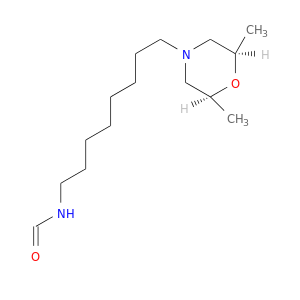 |
| DB04114 | acetylcholinesterase | experimental | 3-Chloro-9-Ethyl-6,7,8,9,10,11-Hexahydro-7,11-Methanocycloocta[B]Quinolin-12-Amine |  | 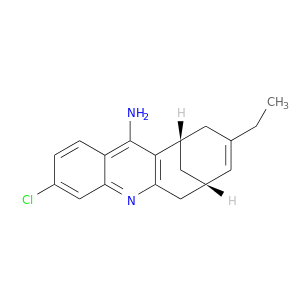 |
| DB04556 | acetylcholinesterase | experimental | 3-[(1s)-1-(Dimethylamino)Ethyl]Phenol |  |  |
| DB04614 | acetylcholinesterase | experimental | (R)-tacrine(10)-hupyridone |  |  |
| DB04615 | acetylcholinesterase | experimental | (S)-tacrine(10)-hupyridone |  |  |
| DB04616 | acetylcholinesterase | experimental | TACRINE(8)-4-AMINOQUINOLINE | 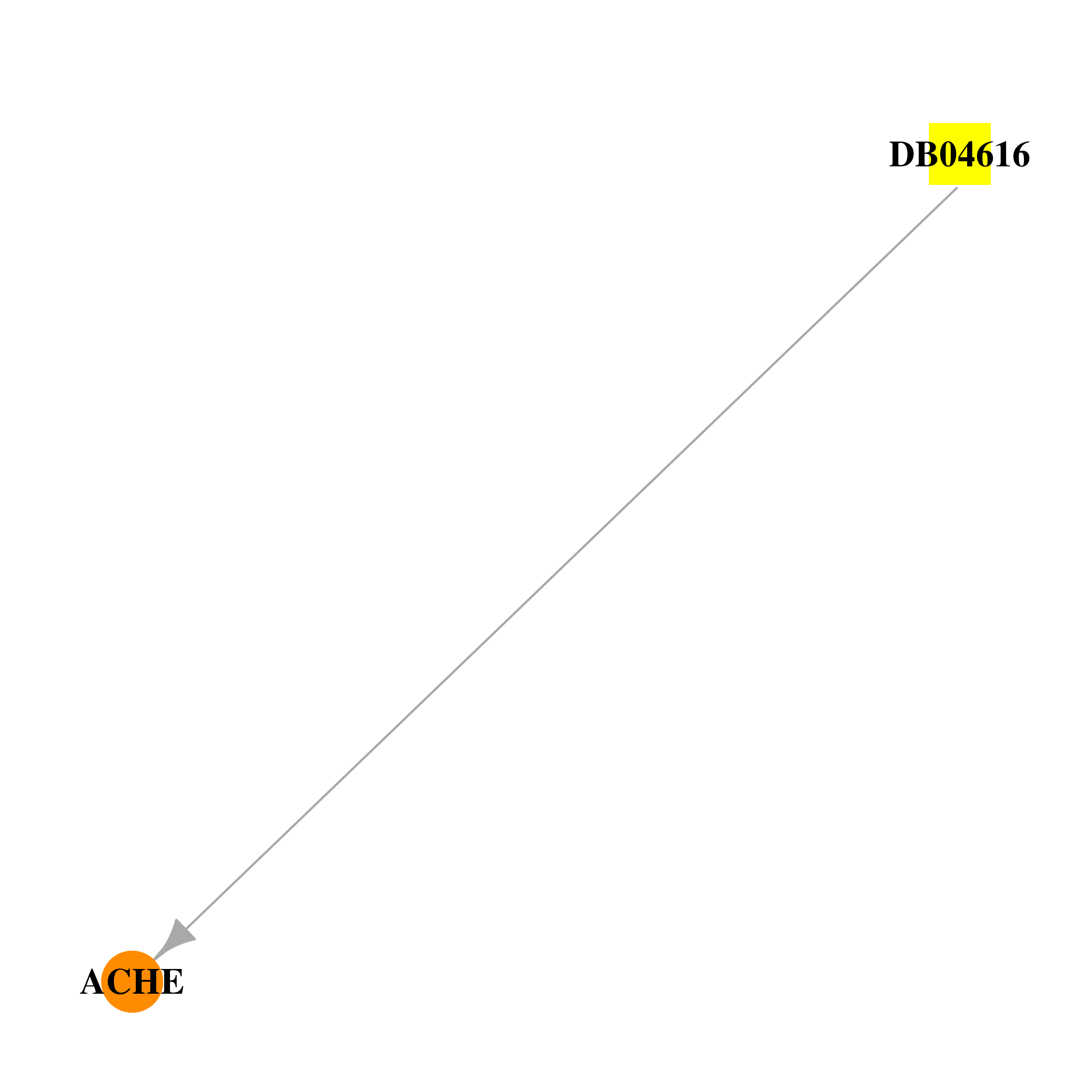 | 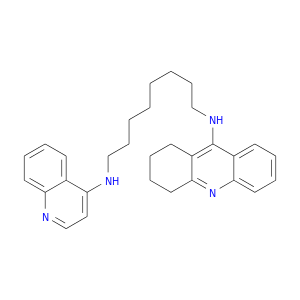 |
| DB04617 | acetylcholinesterase | experimental | (9S)-9-[(8-AMMONIOOCTYL)AMINO]-1,2,3,4,9,10-HEXAHYDROACRIDINIUM |  |  |
| DB07555 | acetylcholinesterase | experimental | 1-(2-nitrophenyl)-2,2,2-trifluoroethyl]-arsenocholine |  |  |
| DB07701 | acetylcholinesterase | experimental | 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE |  |  |
| DB07756 | acetylcholinesterase | experimental | 1-[3-({[(4-AMINO-5-FLUORO-2-METHYLQUINOLIN-3-YL)METHYL]THIO}METHYL)PHENYL]-2,2,2-TRIFLUOROETHANE-1,1-DIOL |  | 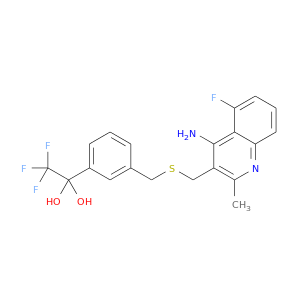 |
| DB07822 | acetylcholinesterase | experimental | (1R)-1,2,2-TRIMETHYLPROPYL (S)-METHYLPHOSPHINATE |  | 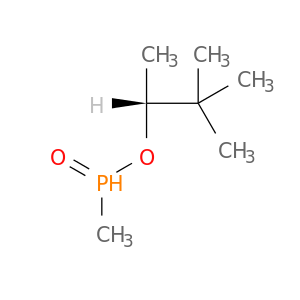 |
| DB07846 | acetylcholinesterase | experimental | 1S,3AS,8AS-TRIMETHYL-1-OXIDO-1,2,3,3A,8,8A-HEXAHYDROPYRROLO[2,3-B]INDOL-5-YL 2-ETHYLPHENYLCARBAMATE |  |  |
| DB08167 | acetylcholinesterase | experimental | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM |  |  |
| DB08357 | acetylcholinesterase | experimental | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE |  |  |
| DB08615 | acetylcholinesterase | experimental | 2-[4-(DIMETHYLAMINO)PHENYL]-6-HYDROXY-3-METHYL-1,3-BENZOTHIAZOL-3-IUM |  |  |
| Top |
| Cross referenced IDs for ACHE |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |