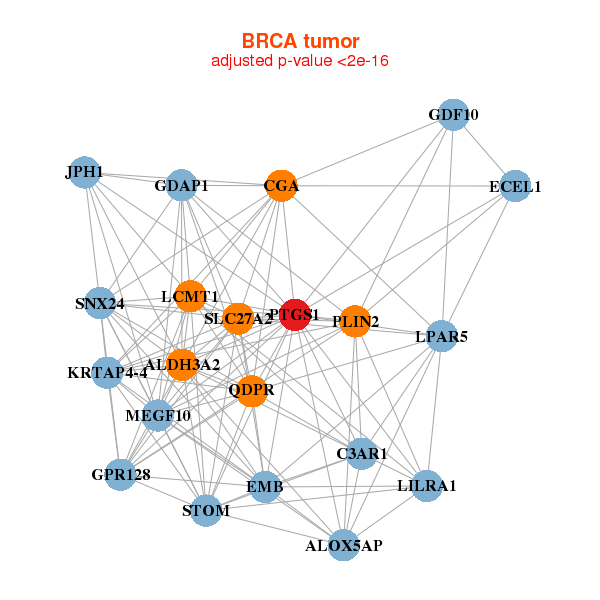
ALDH3A2,ALOX5AP,C3AR1,CGA,ECEL1,EMB,GDAP1,
GDF10,GPR128,JPH1,KRTAP4-4,LCMT1,LILRA1,LPAR5,
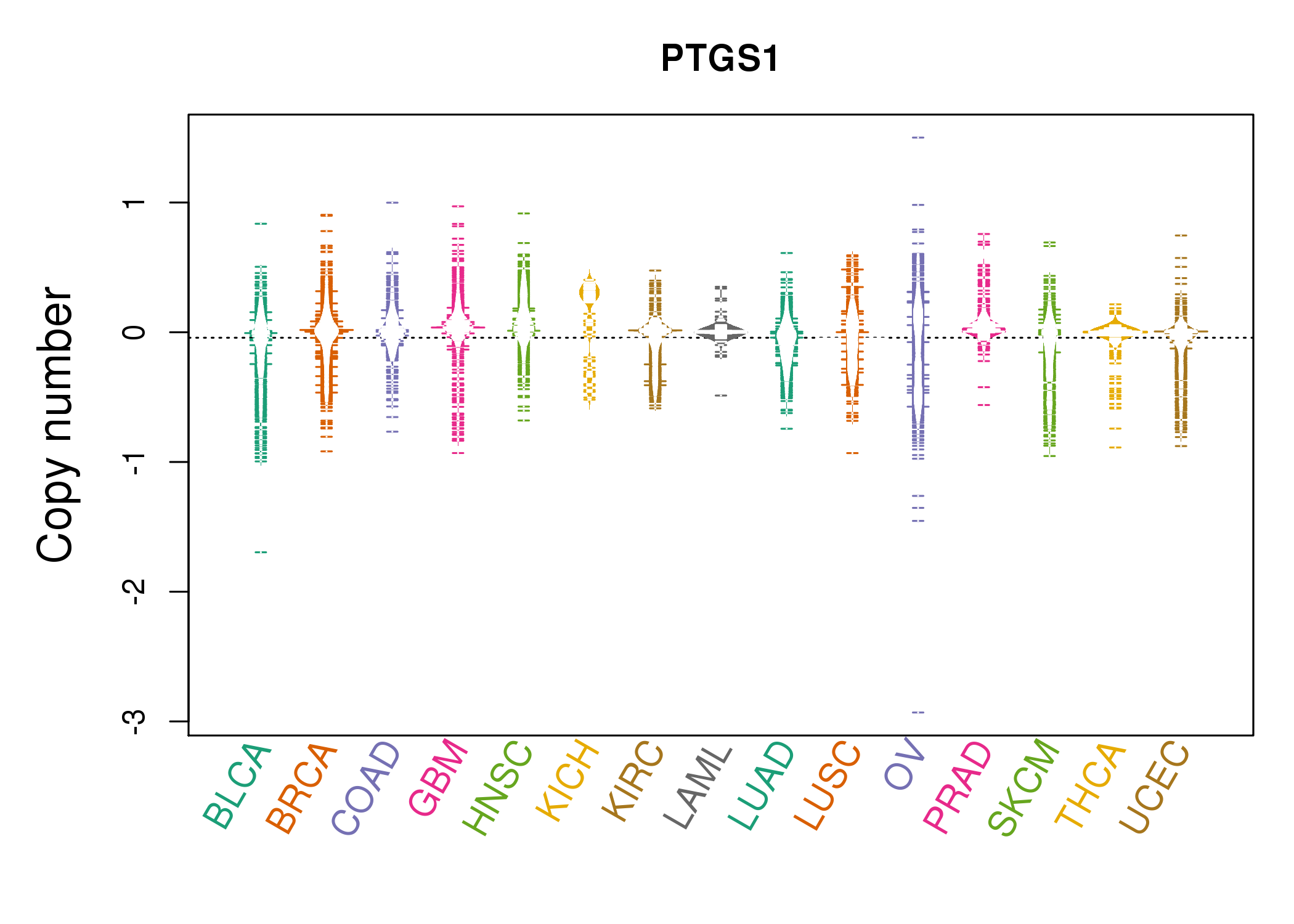
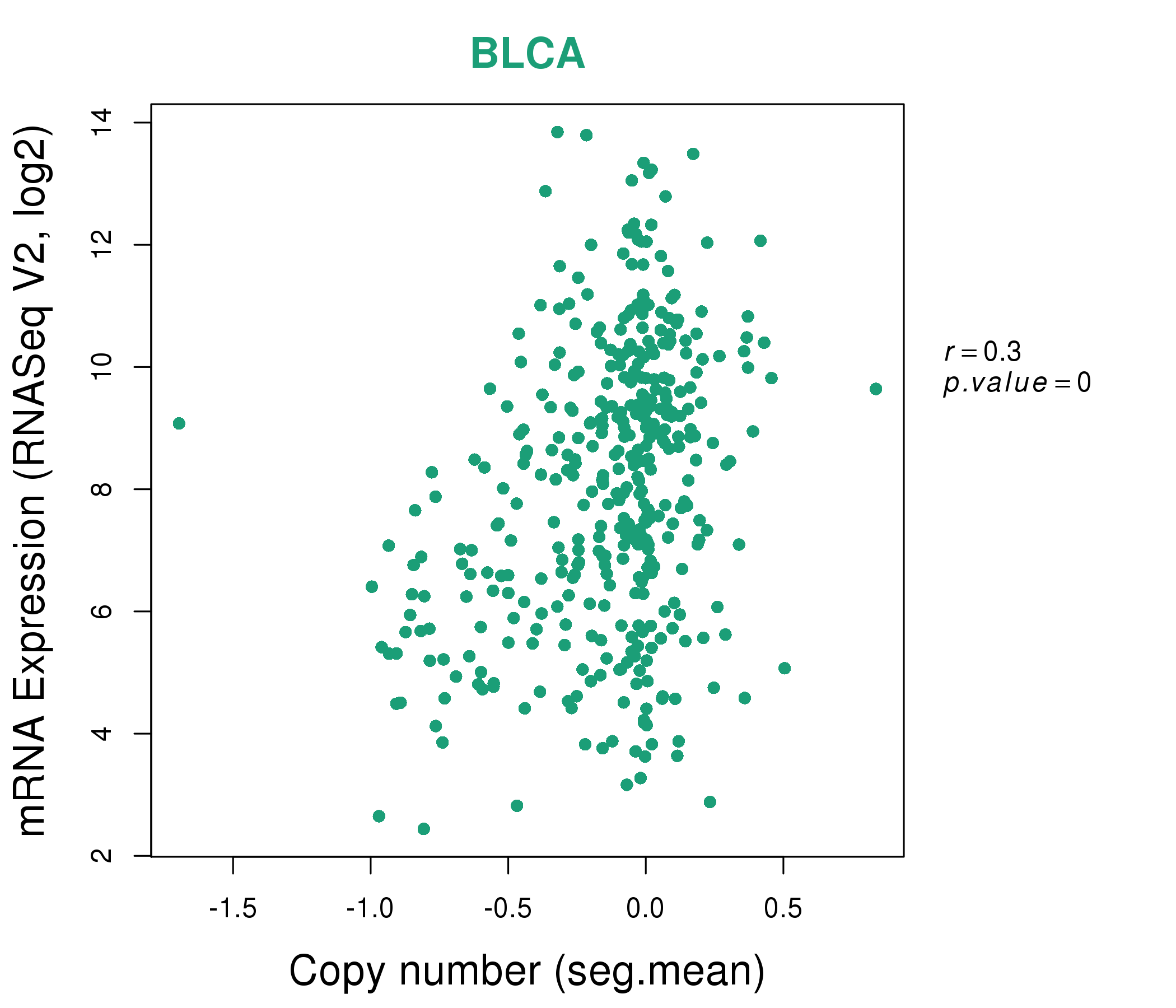
MEGF10,PLIN2,PTGS1,QDPR,SLC27A2,SNX24,STOM | AHNAK2,ALOX5AP,AXL,C1S,CSF1R,CYBB,DNM1,
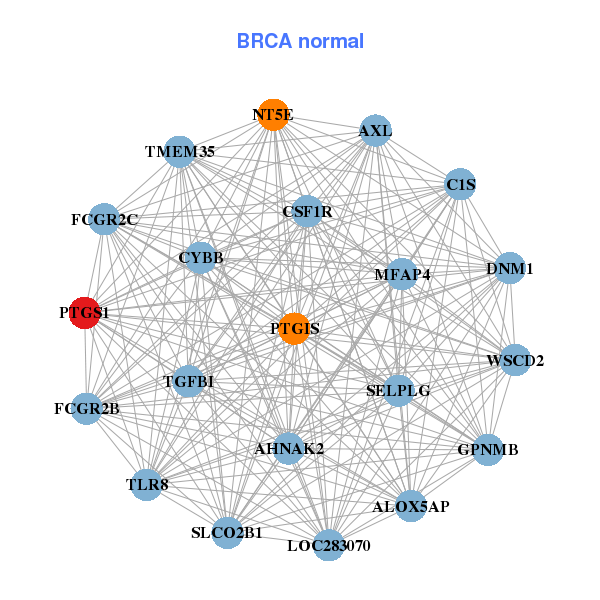
FCGR2B,FCGR2C,GPNMB,LOC283070,MFAP4,NT5E,PTGIS,
PTGS1,SELPLG,SLCO2B1,TGFBI,TLR8,TMEM35,WSCD2 |
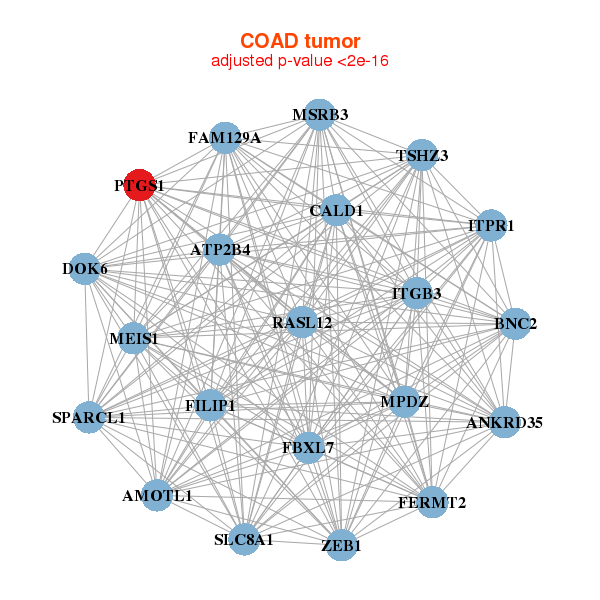
AMOTL1,ANKRD35,ATP2B4,BNC2,CALD1,DOK6,FAM129A,
FBXL7,FERMT2,FILIP1,ITGB3,ITPR1,MEIS1,MPDZ,
MSRB3,PTGS1,RASL12,SLC8A1,SPARCL1,TSHZ3,ZEB1 | ATP2B4,CACNA1C,CACNA1H,DMD,FRMPD4,GNAO1,KCNH2,
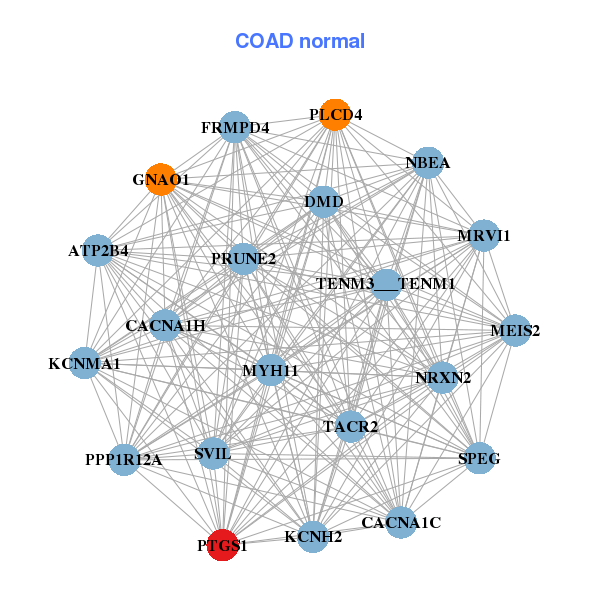
KCNMA1,MEIS2,MRVI1,MYH11,NBEA,NRXN2,TENM3___TENM1,
PLCD4,PPP1R12A,PRUNE2,PTGS1,SPEG,SVIL,TACR2 |
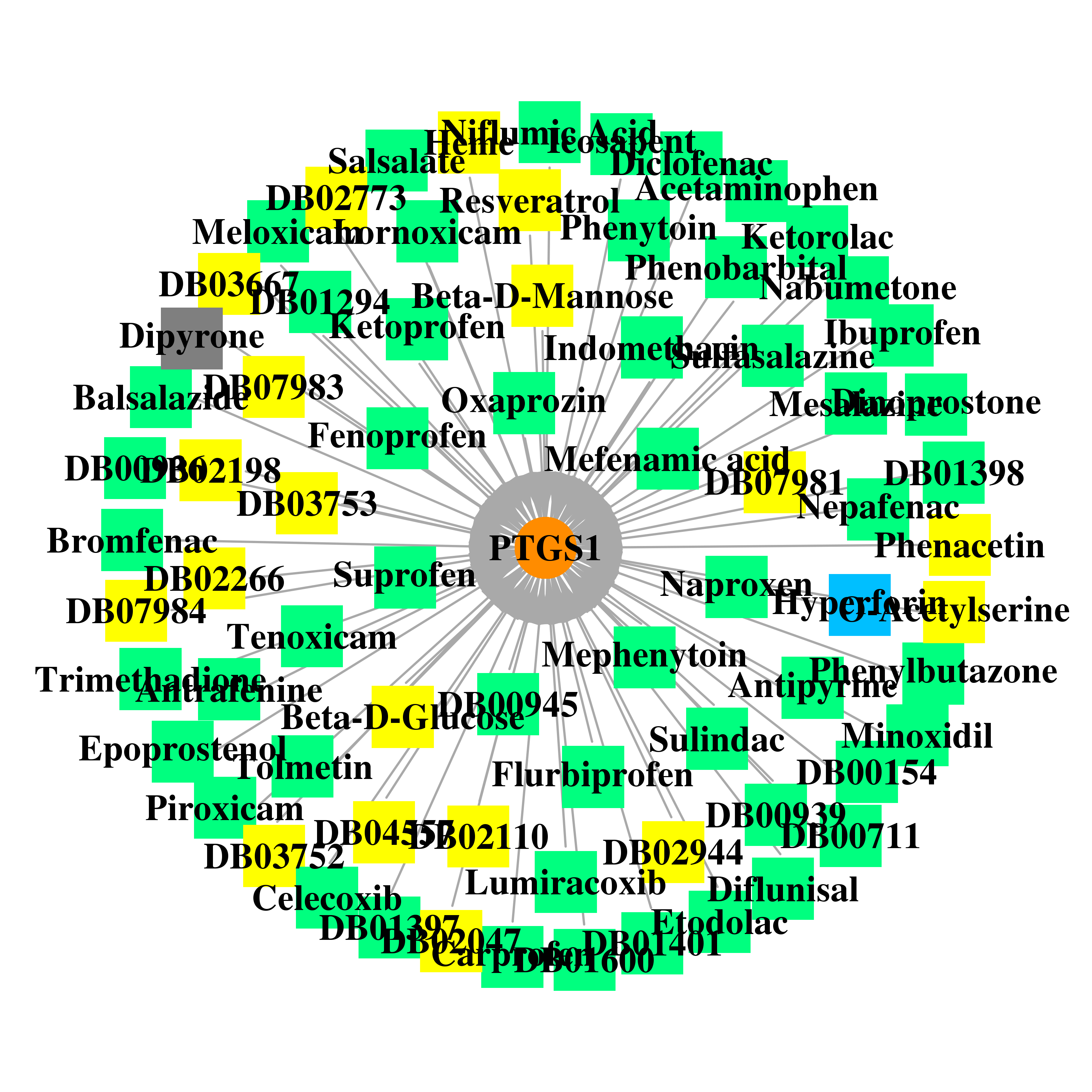
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB00154 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved; nutraceutical | gamma-Homolinolenic acid | 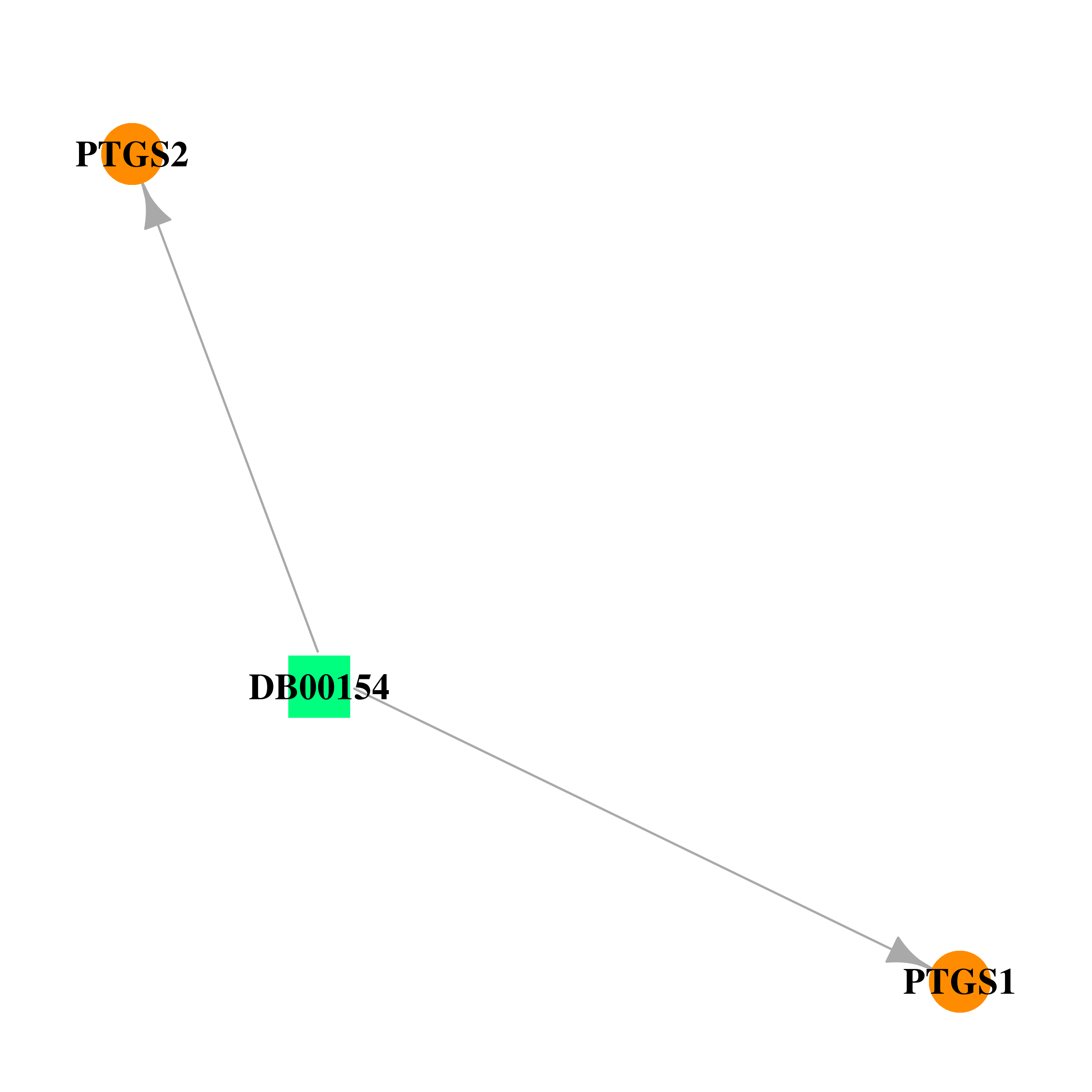 |  |
| DB00159 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved; nutraceutical | Icosapent | 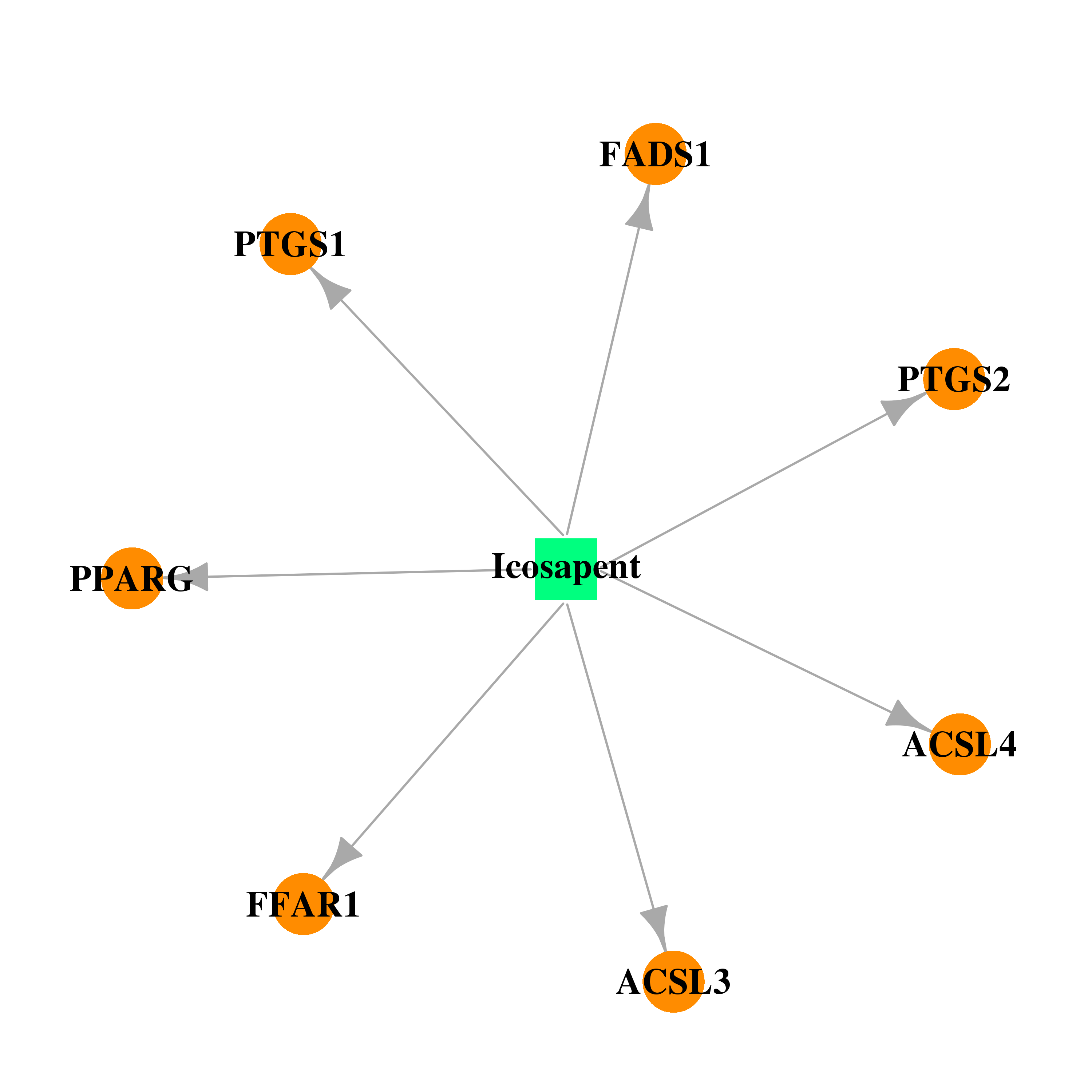 |  |
| DB00244 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Mesalazine | 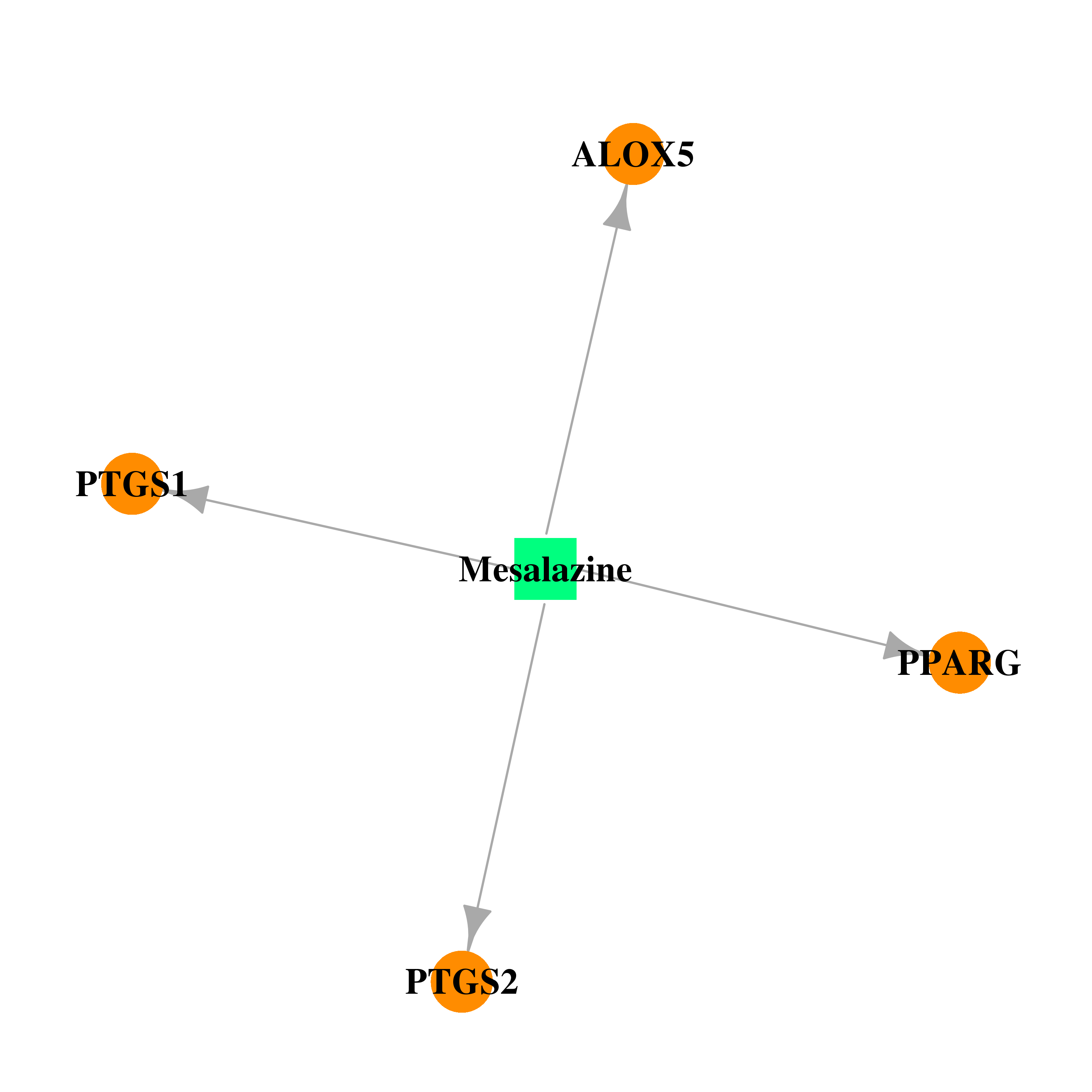 |  |
| DB00316 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Acetaminophen |  |  |
| DB00328 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved; investigational | Indomethacin |  | 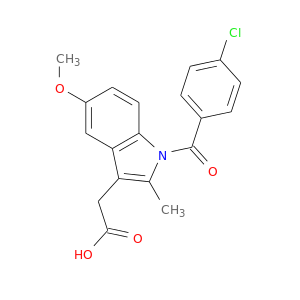 |
| DB00350 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Minoxidil | 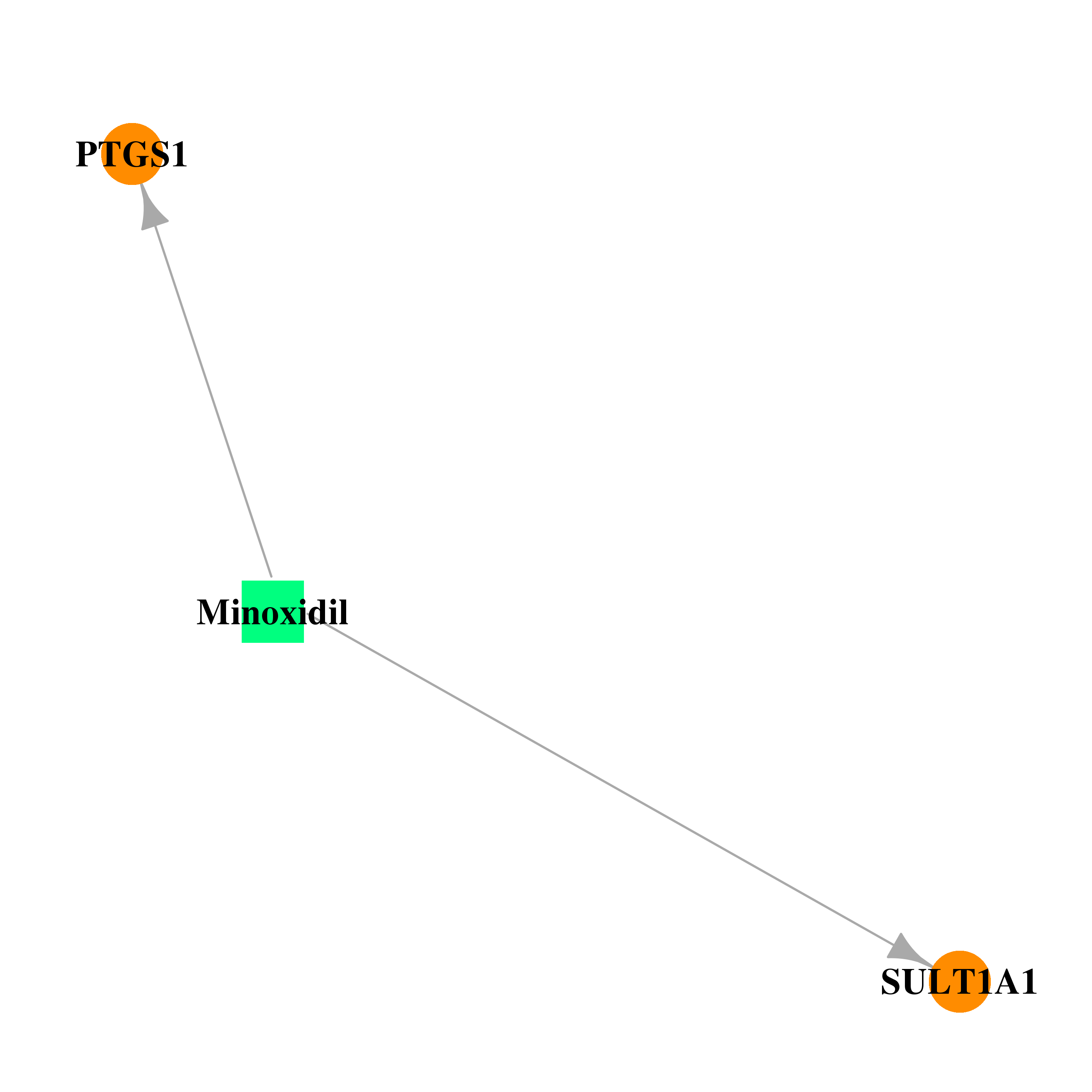 |  |
| DB00461 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Nabumetone | 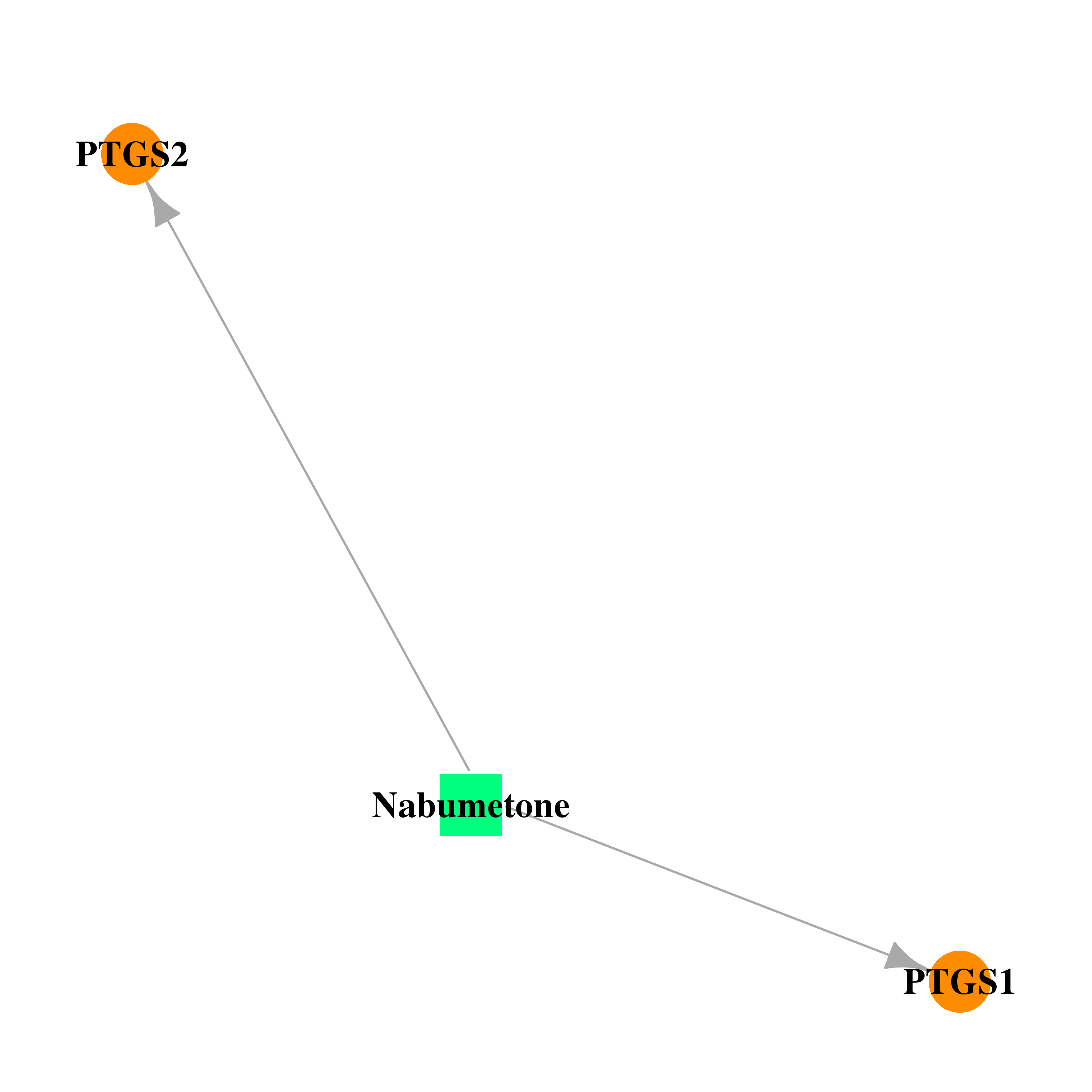 | 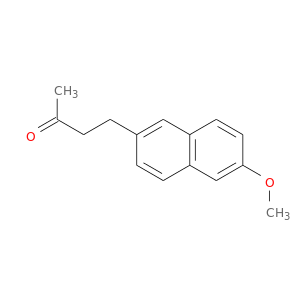 |
| DB00465 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Ketorolac | 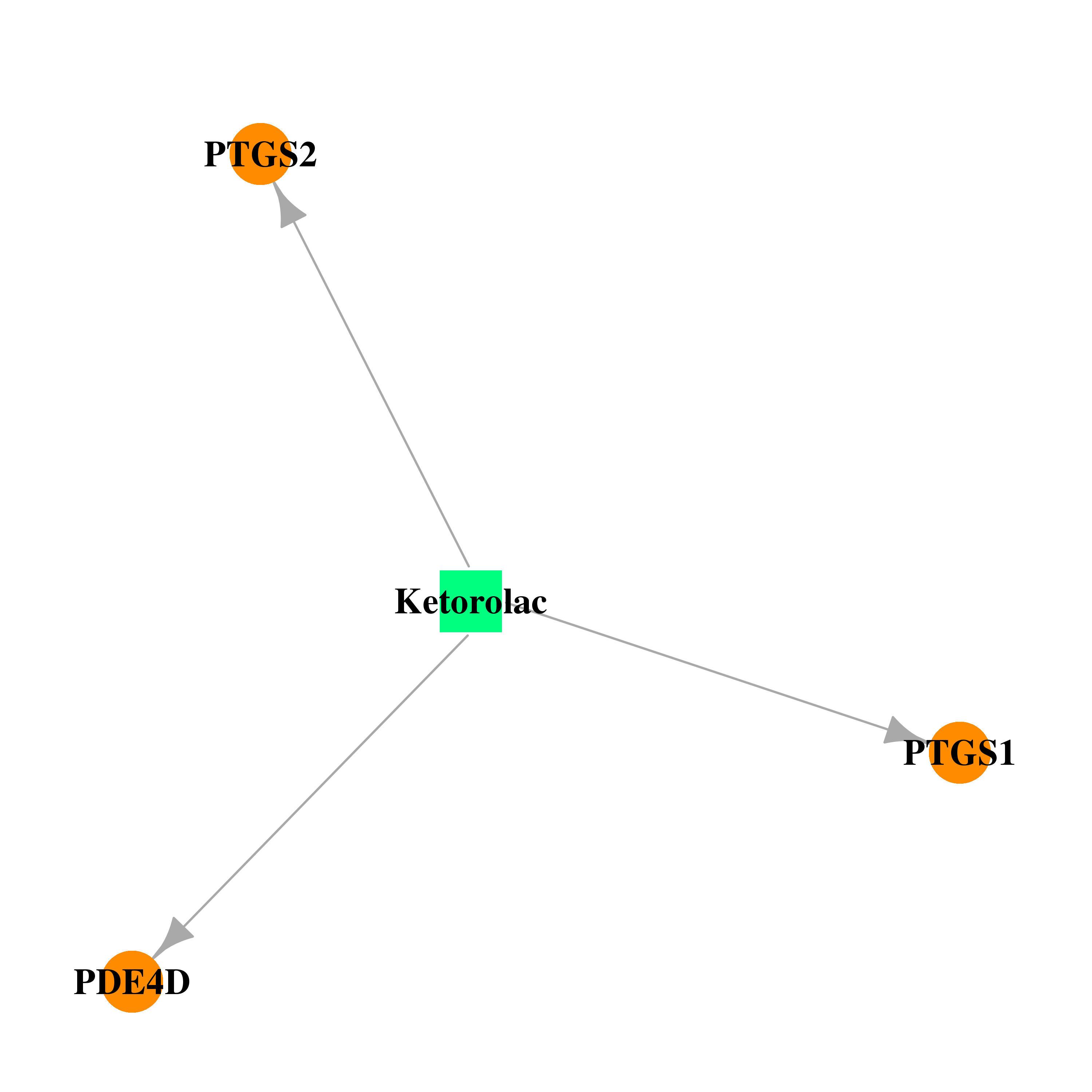 |  |
| DB00469 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Tenoxicam |  |  |
| DB00500 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Tolmetin | 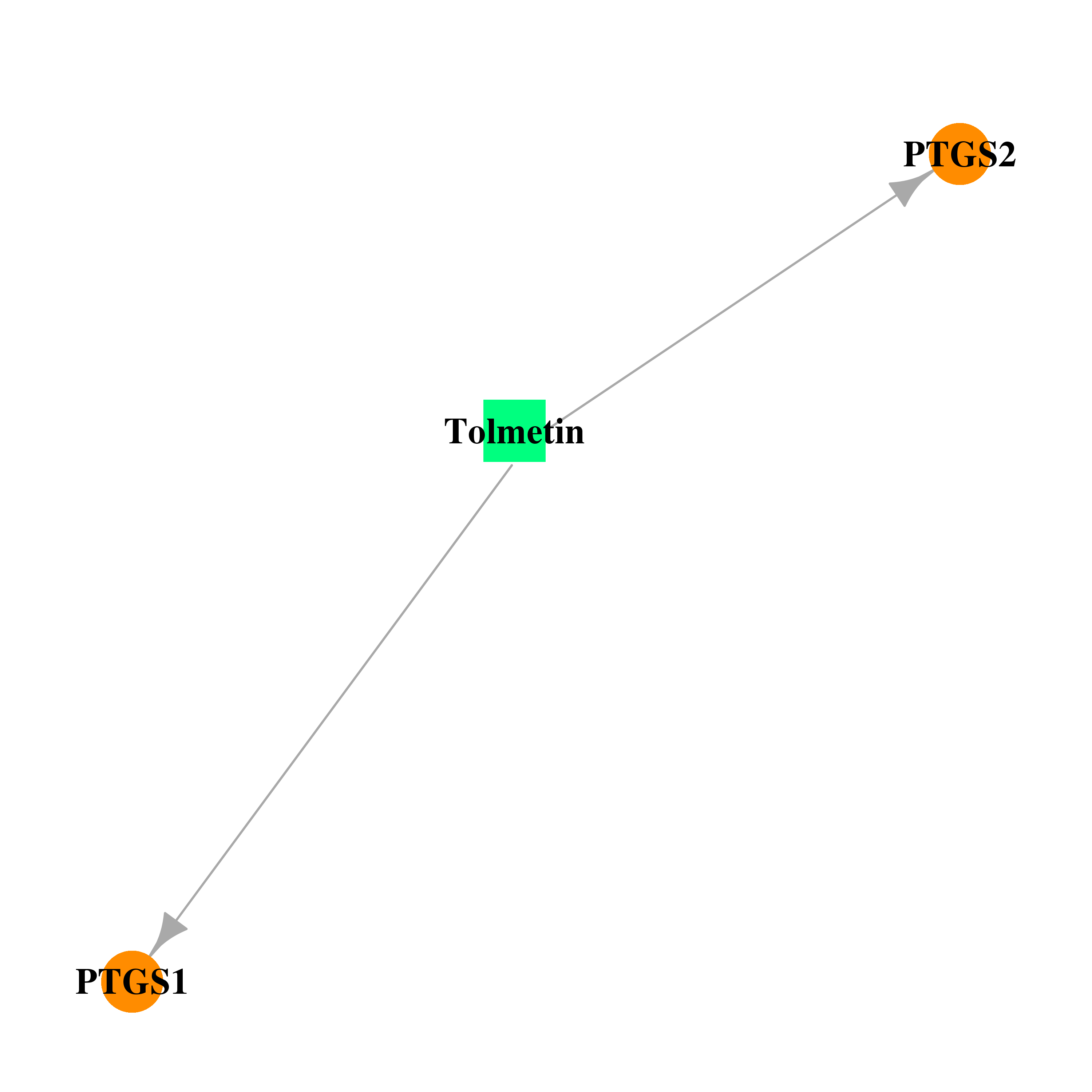 |  |
| DB00554 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved; investigational | Piroxicam | 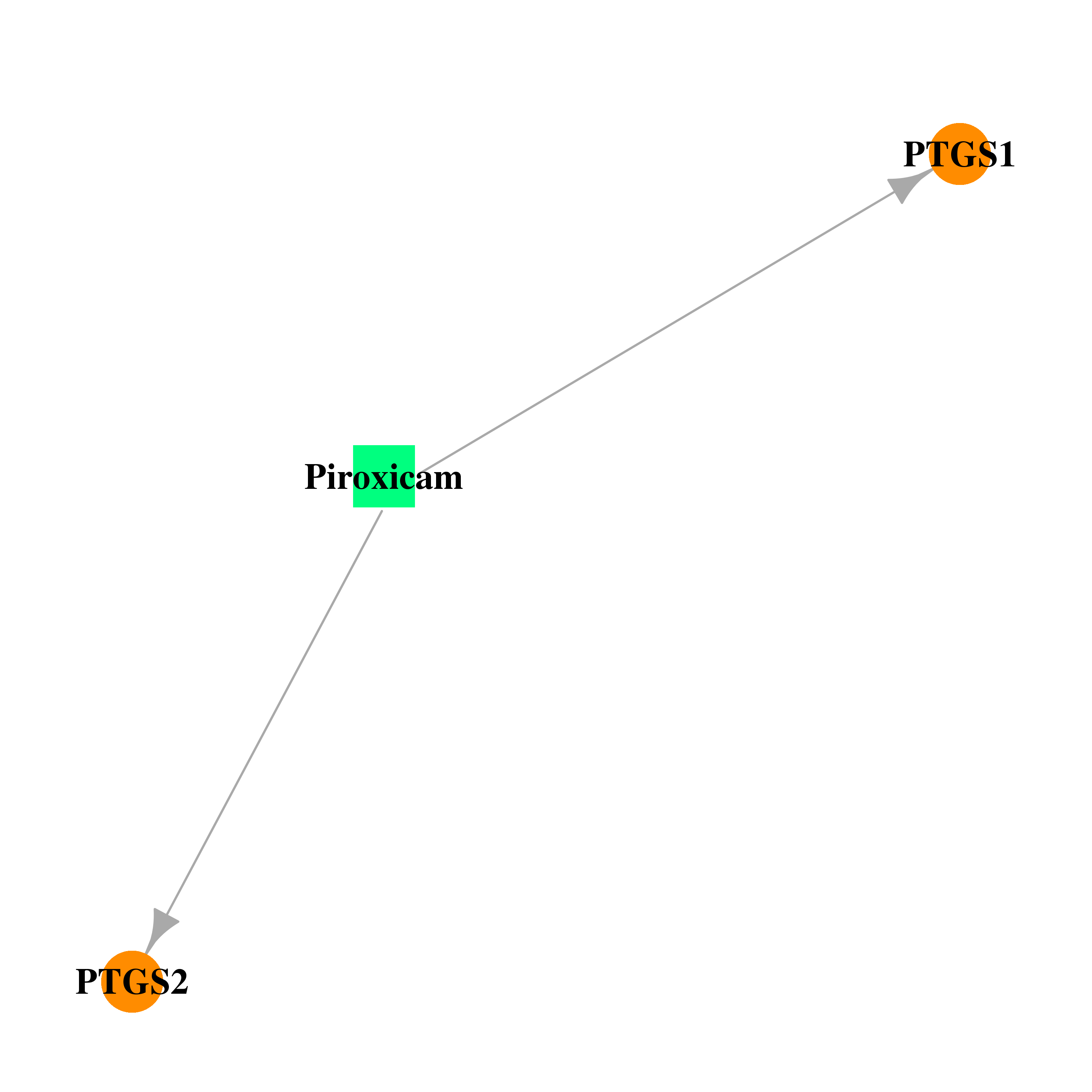 |  |
| DB00573 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Fenoprofen | 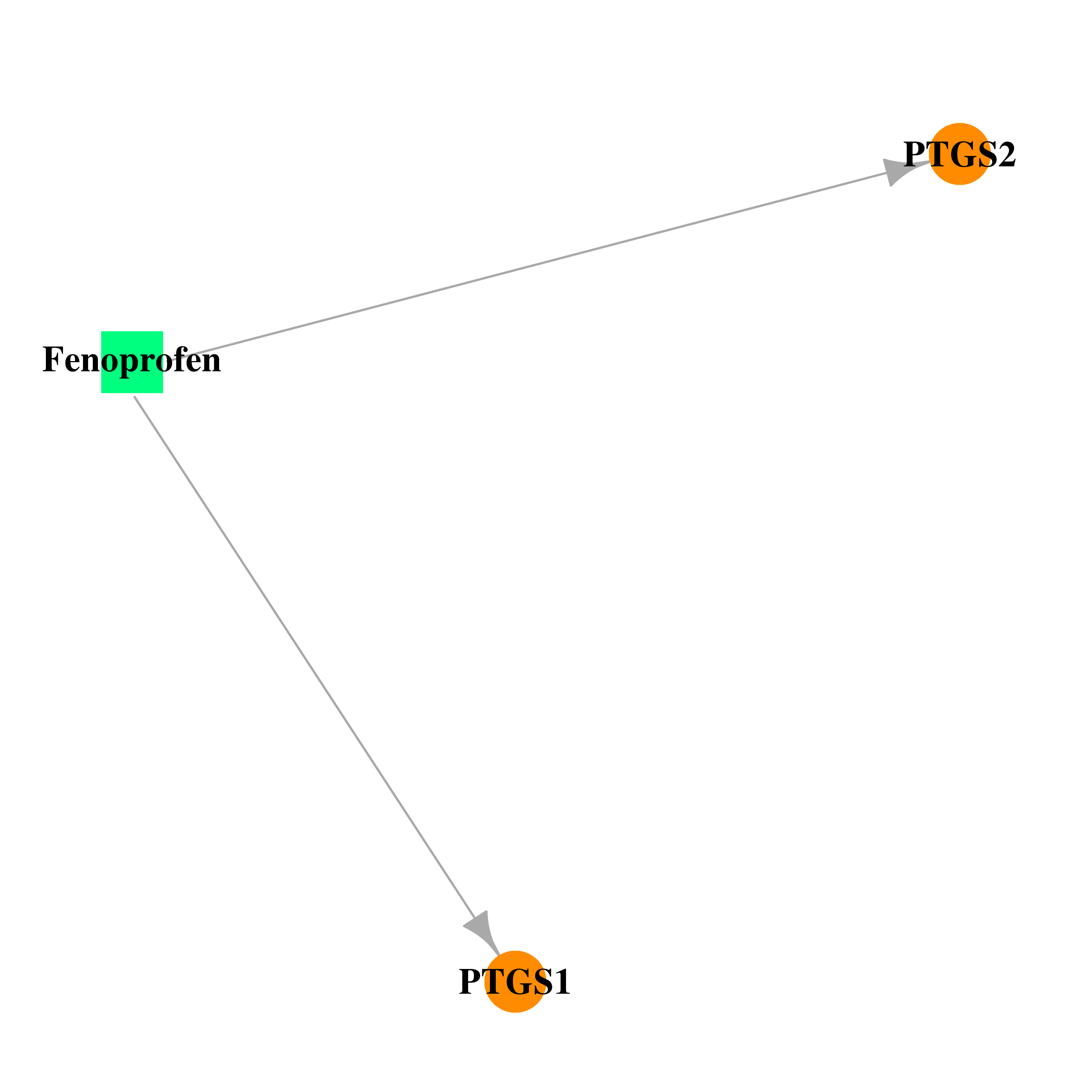 |  |
| DB00586 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Diclofenac |  |  |
| DB00605 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Sulindac |  |  |
| DB00711 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Diethylcarbamazine | 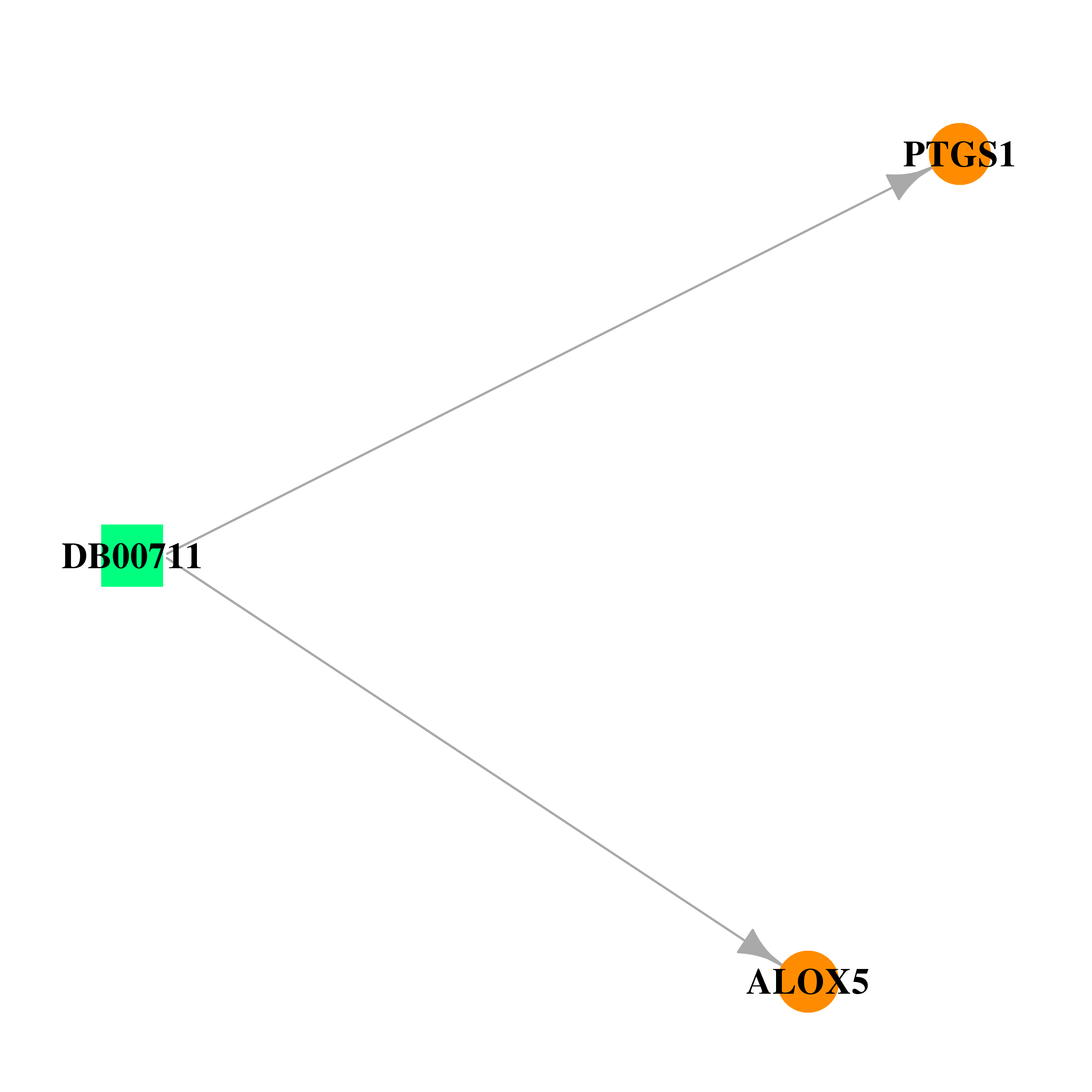 |  |
| DB00712 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Flurbiprofen |  |  |
| DB00749 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved; investigational | Etodolac | 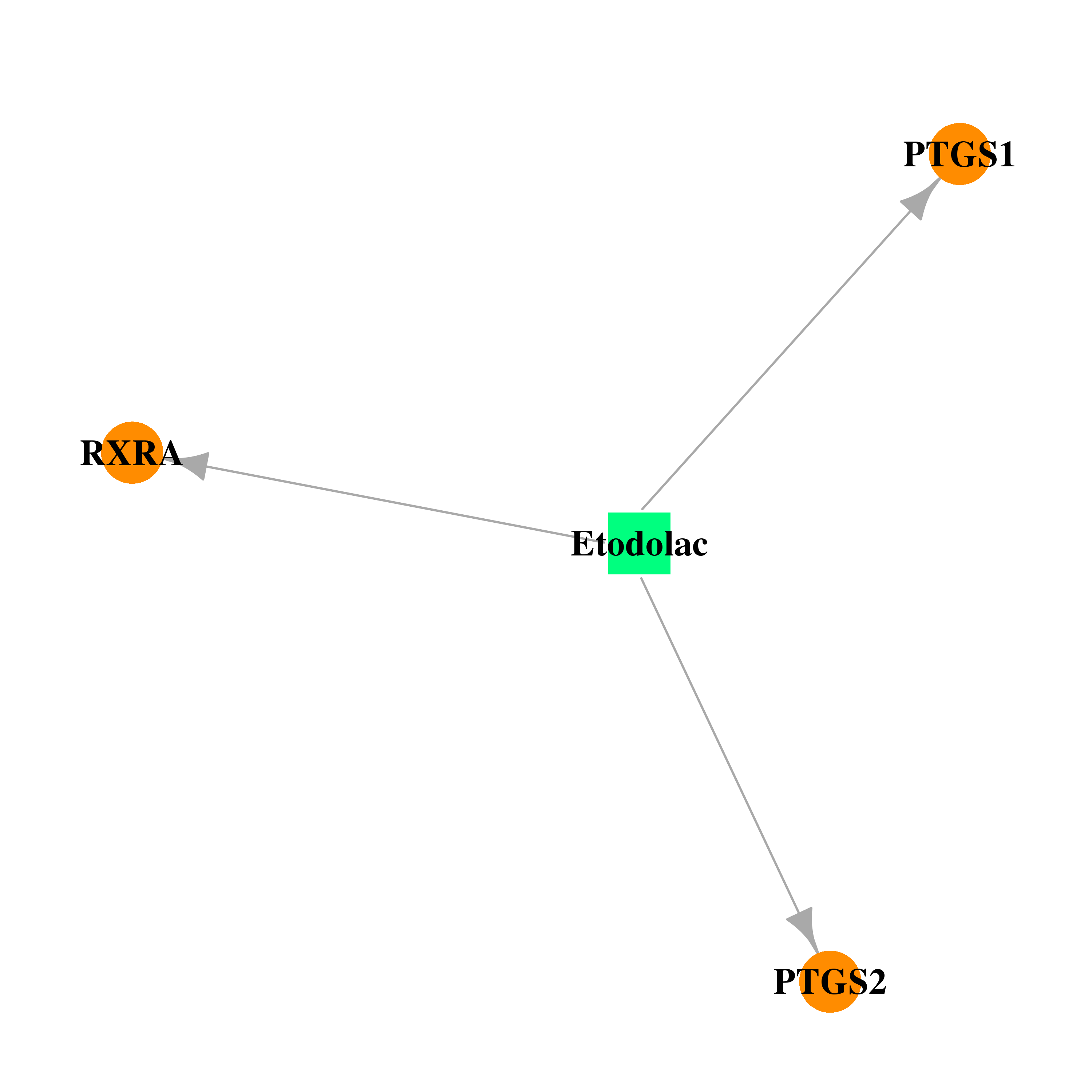 |  |
| DB00784 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Mefenamic acid |  |  |
| DB00788 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Naproxen |  |  |
| DB00795 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Sulfasalazine |  |  |
| DB00812 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Phenylbutazone |  |  |
| DB00814 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Meloxicam |  |  |
| DB00821 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Carprofen |  |  |
| DB00861 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Diflunisal |  |  |
| DB00870 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Suprofen |  |  |
| DB00936 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Salicyclic acid | 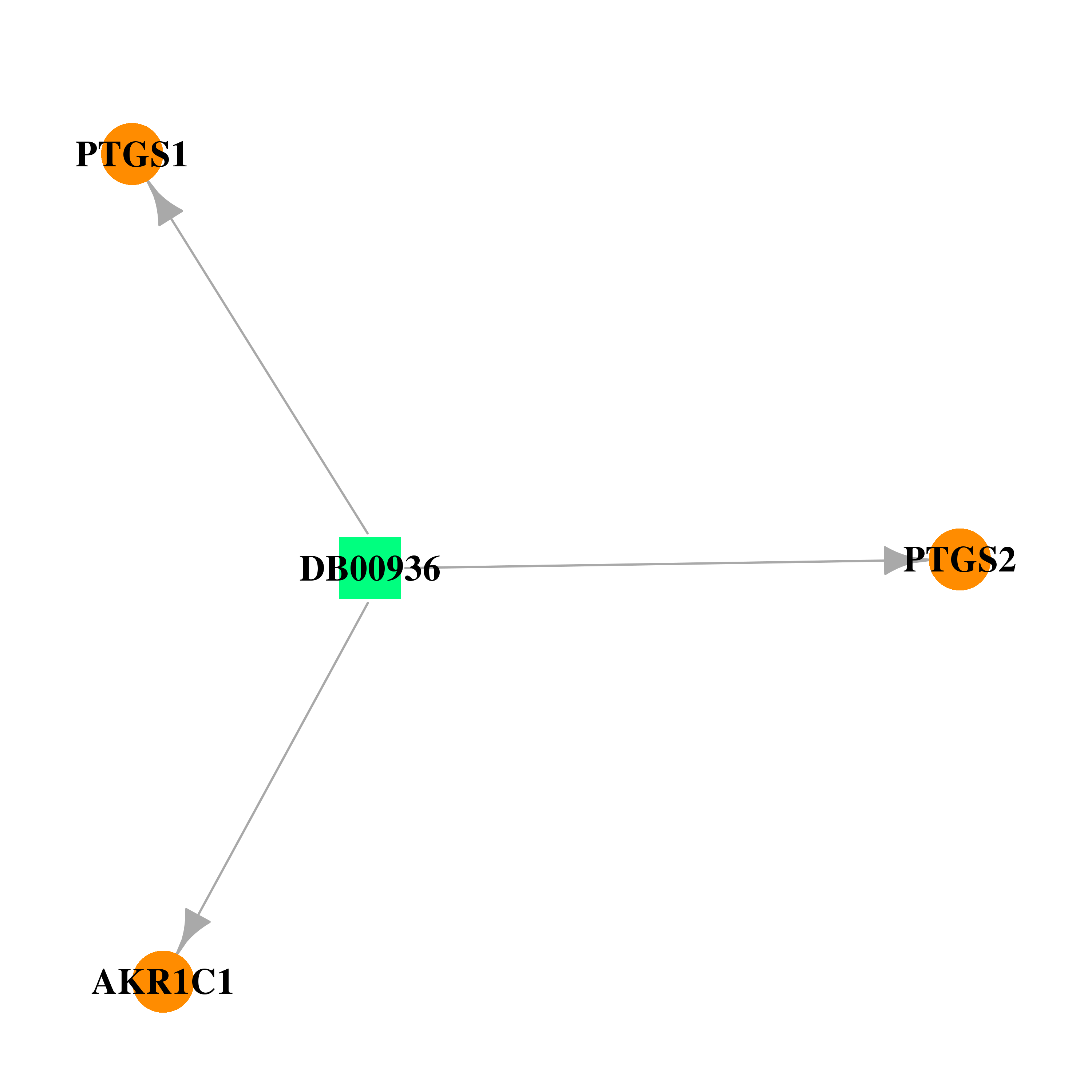 | 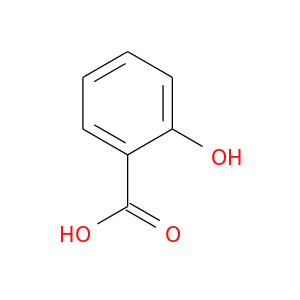 |
| DB00939 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Meclofenamic acid |  |  |
| DB00945 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Acetylsalicylic acid | 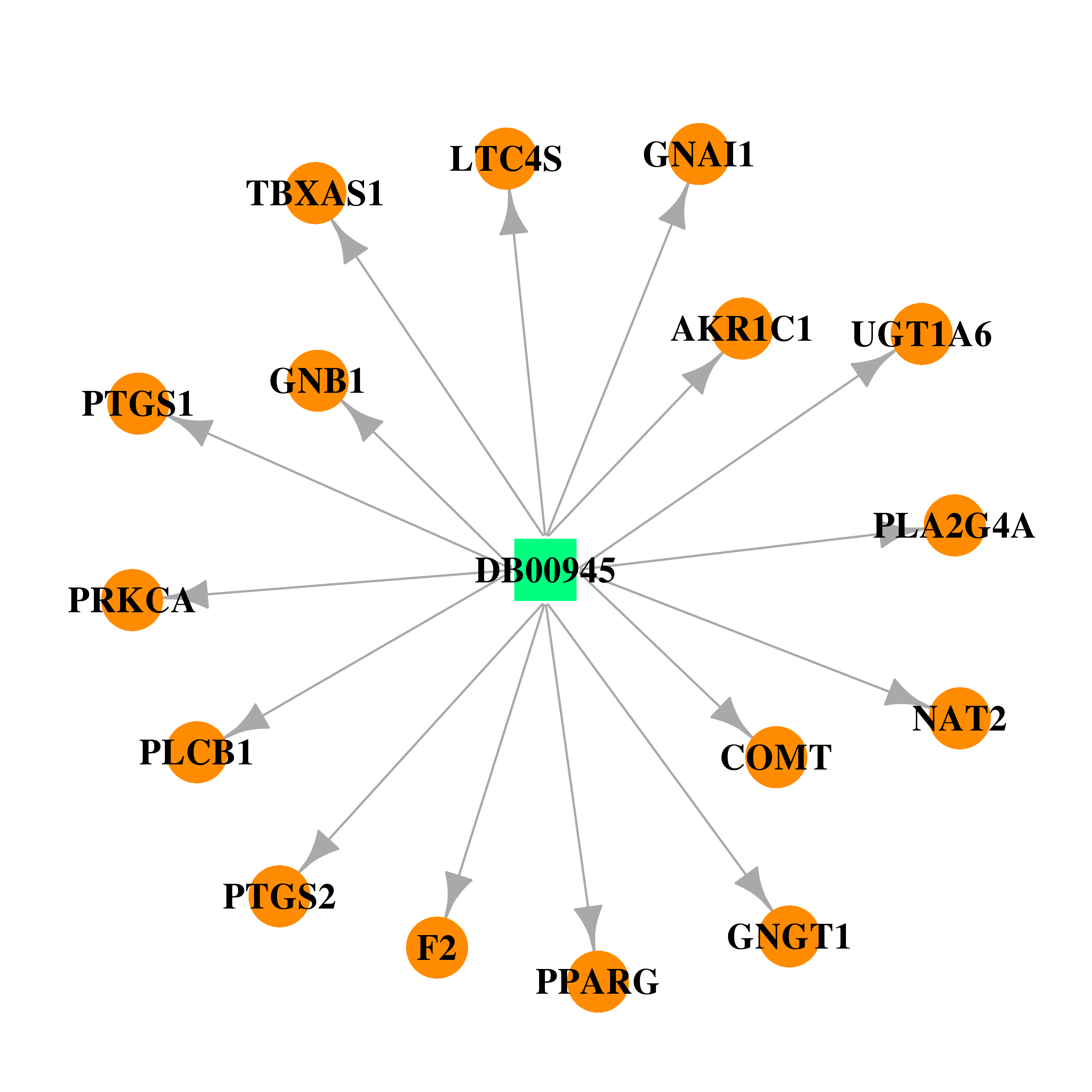 | 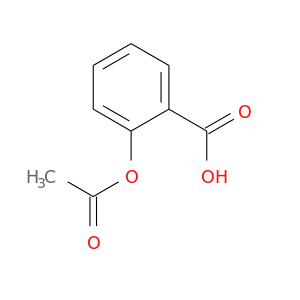 |
| DB00963 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Bromfenac | 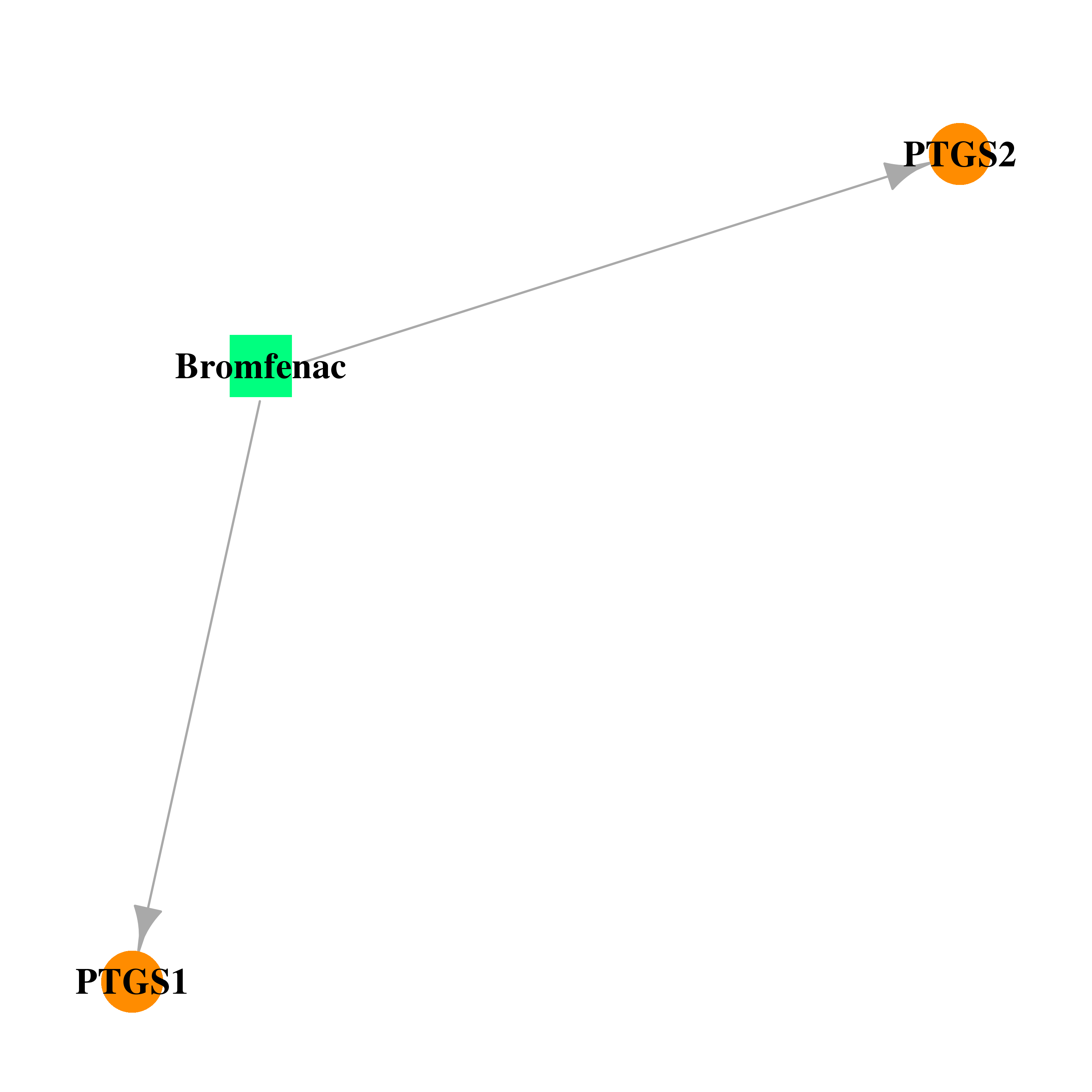 | 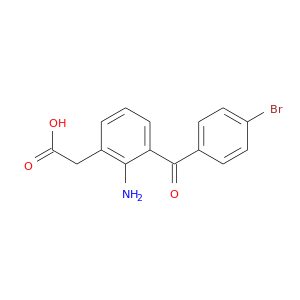 |
| DB00991 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Oxaprozin | 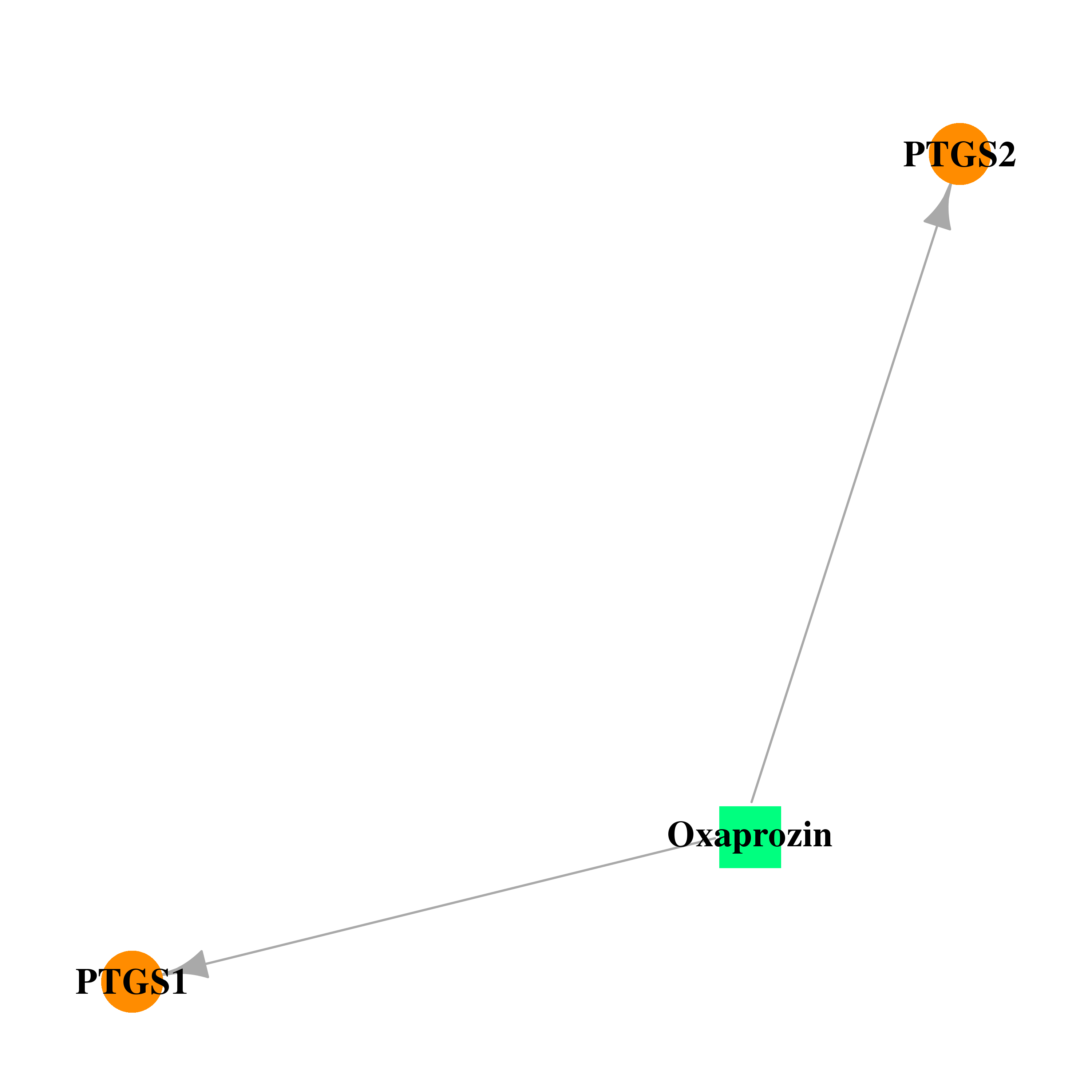 | 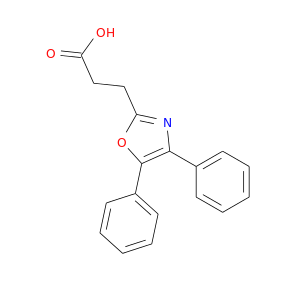 |
| DB01009 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Ketoprofen | 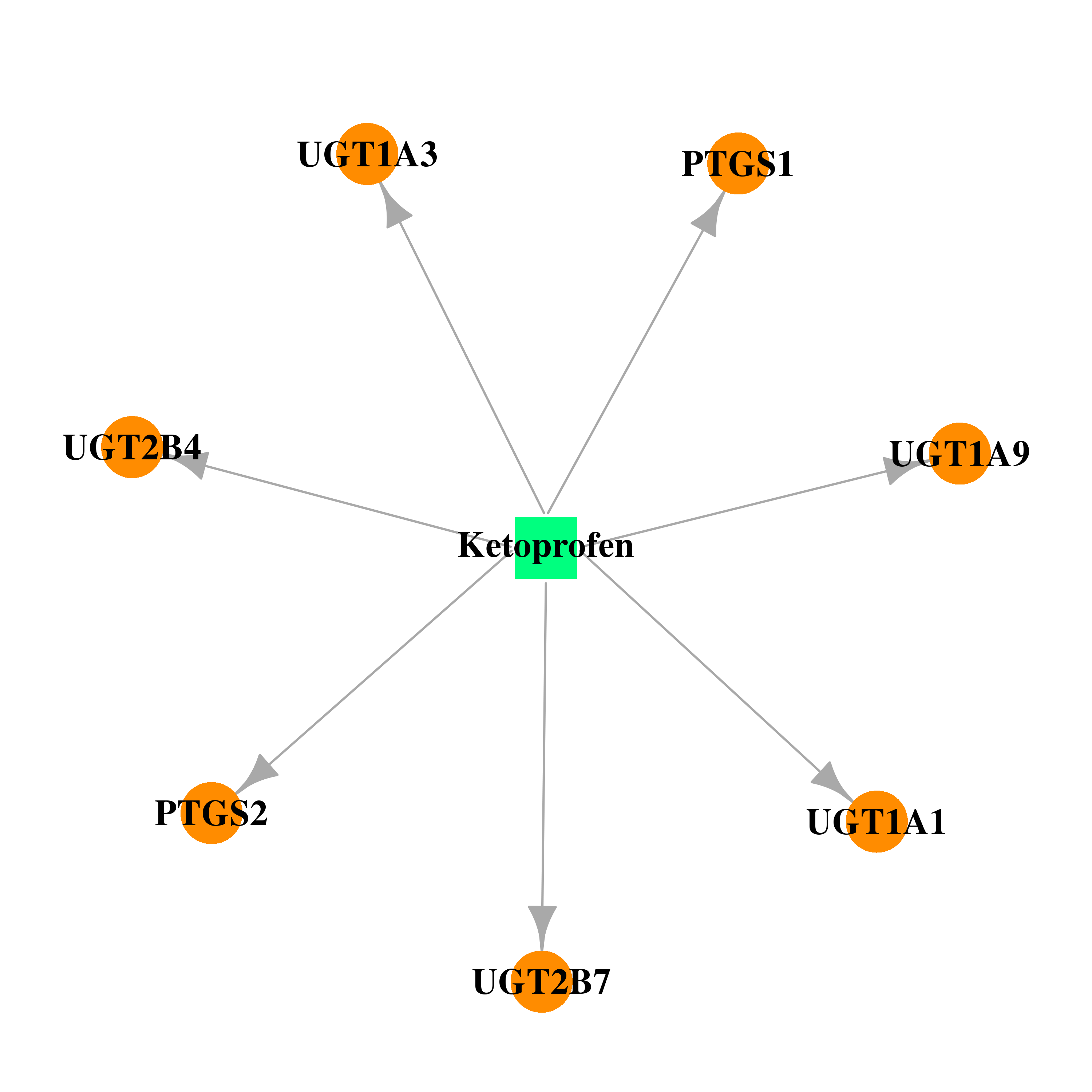 | 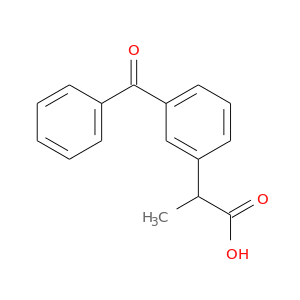 |
| DB01014 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved; investigational | Balsalazide | 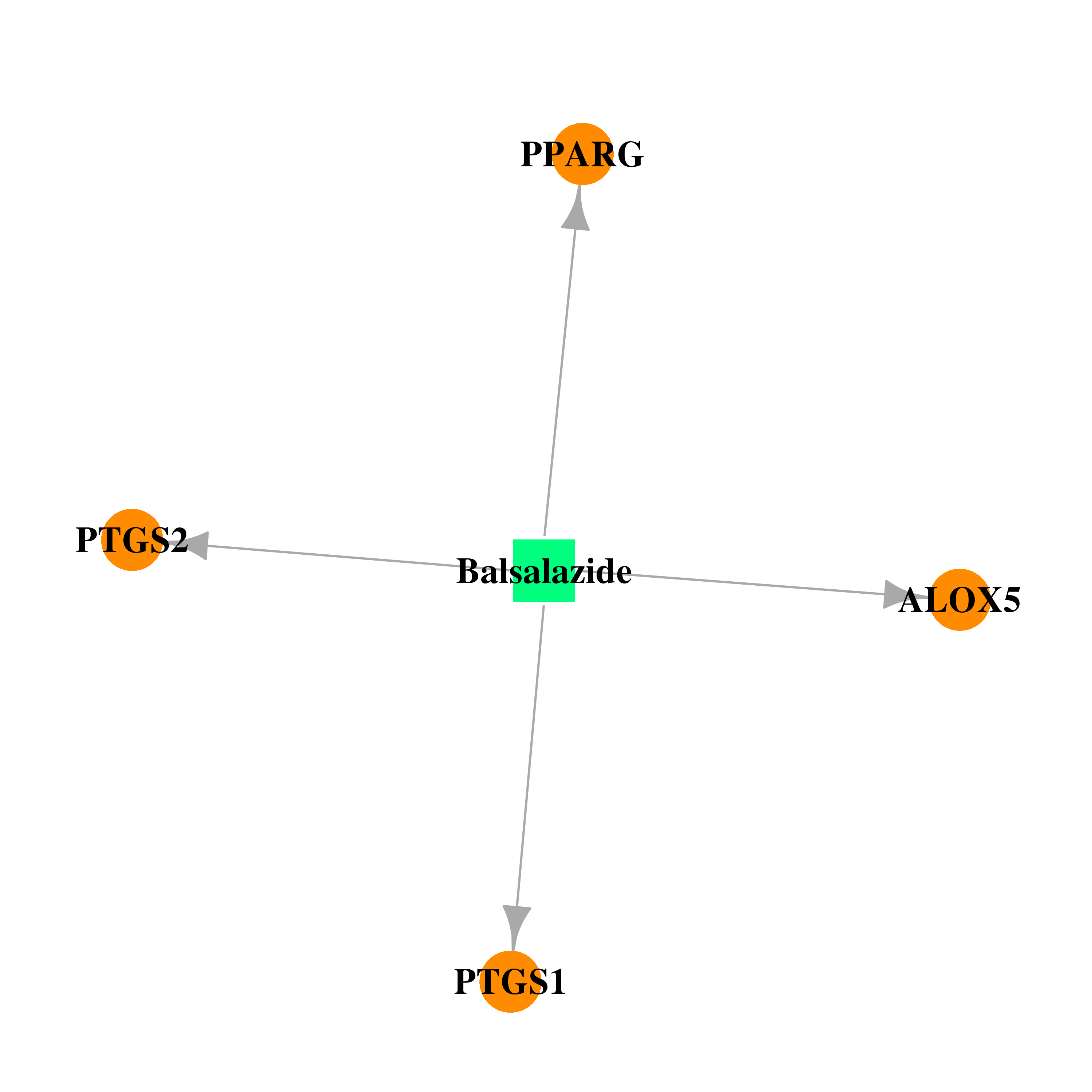 | 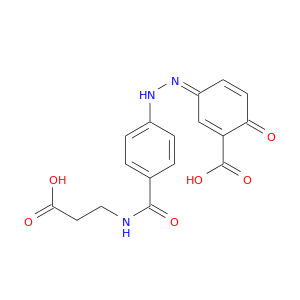 |
| DB01050 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Ibuprofen | 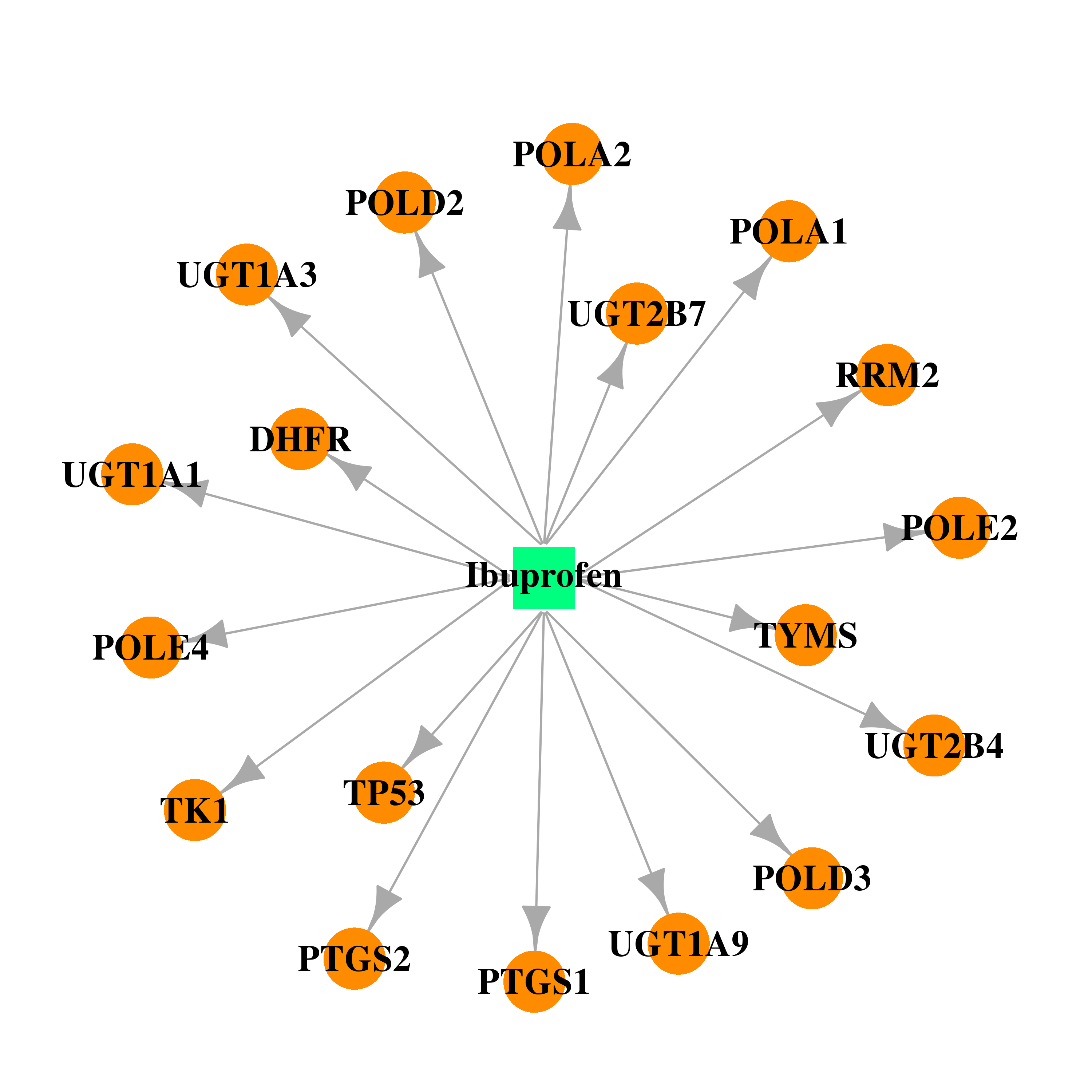 | 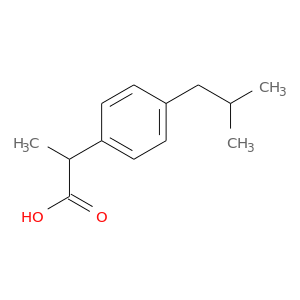 |
| DB01283 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved; investigational | Lumiracoxib |  |  |
| DB01294 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Bismuth Subsalicylate | 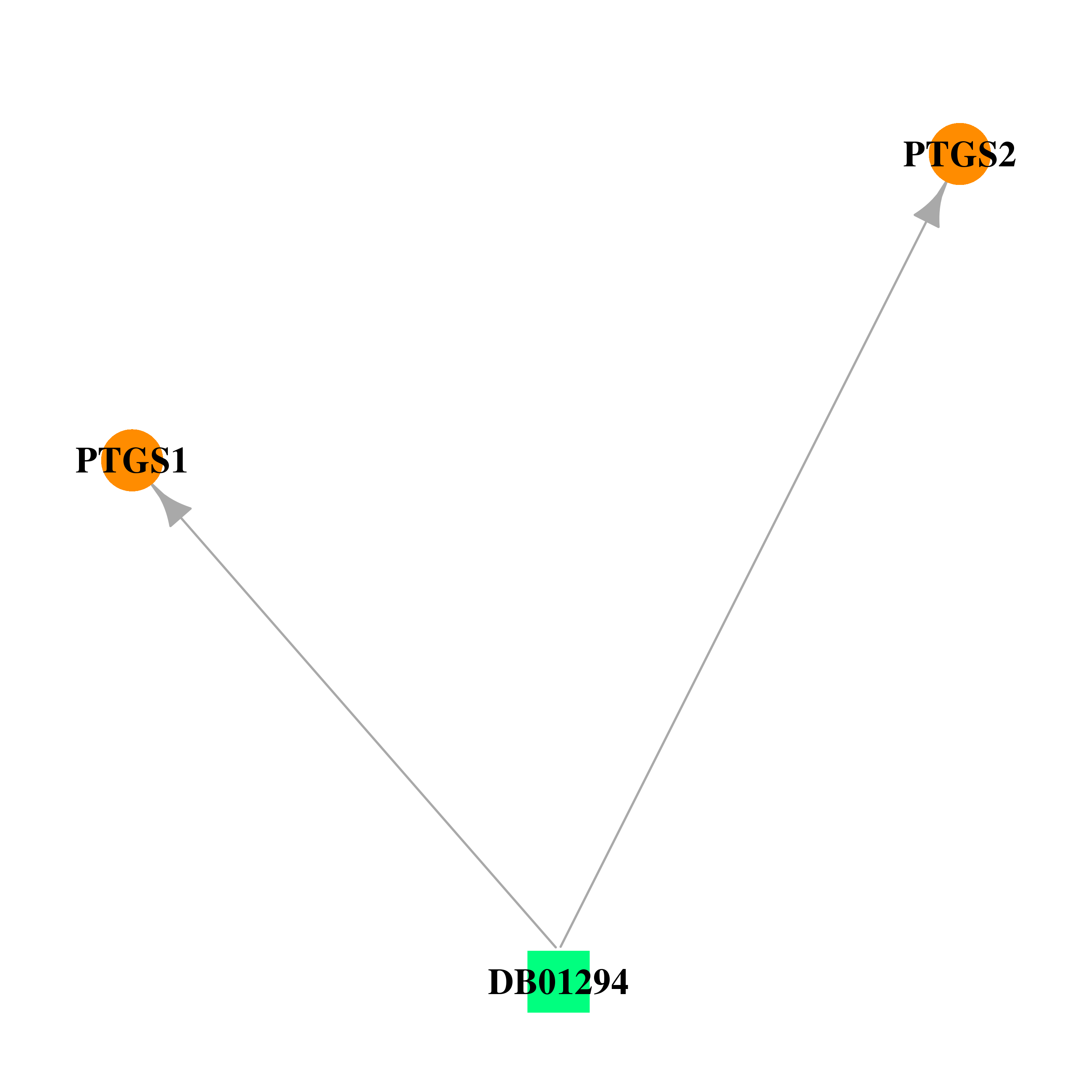 | 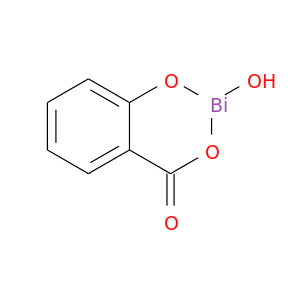 |
| DB01397 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Magnesium salicylate |  |  |
| DB01398 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Salicylate-sodium | 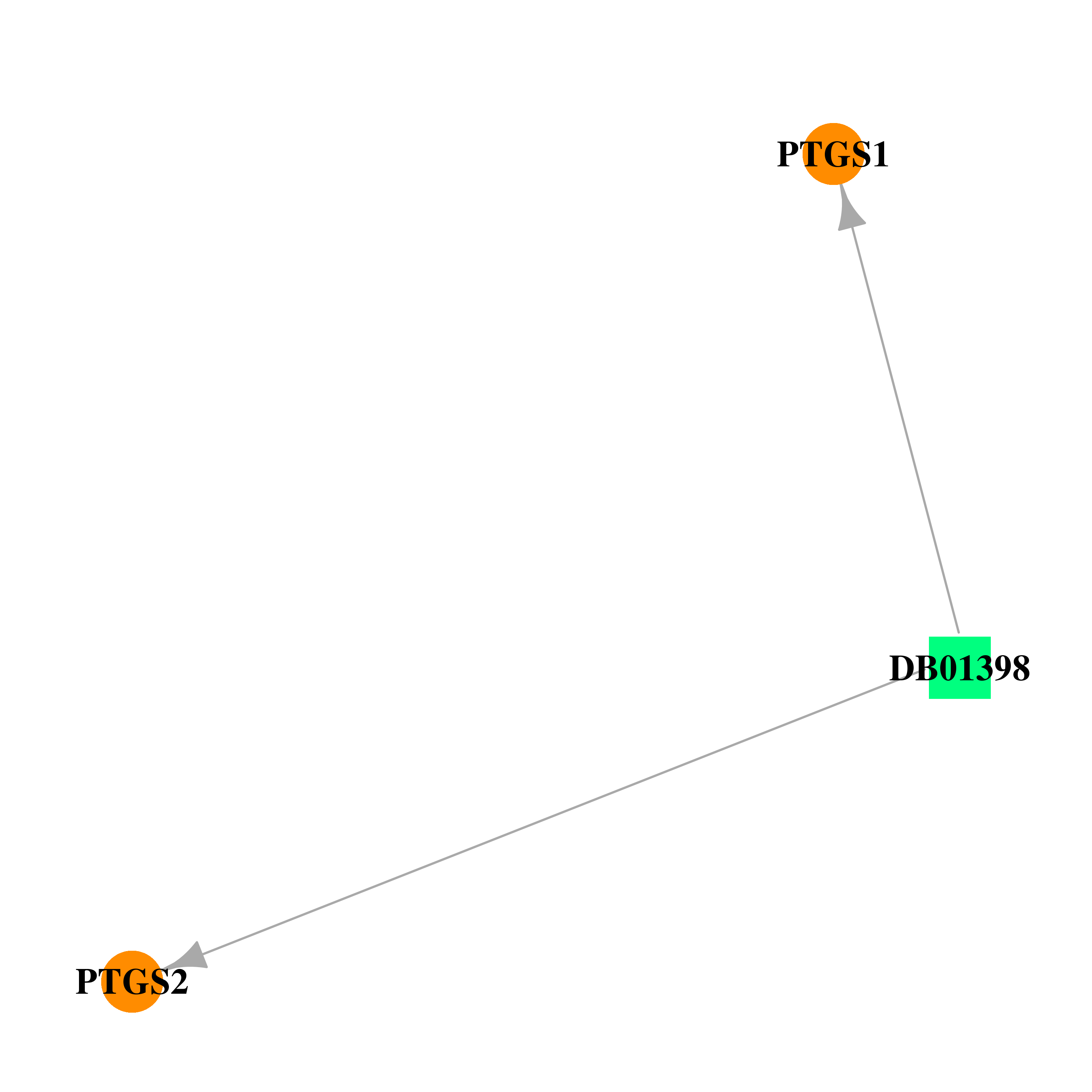 |  |
| DB01399 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Salsalate |  |  |
| DB01401 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Trisalicylate-choline |  |  |
| DB01419 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Antrafenine |  |  |
| DB01435 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Antipyrine | 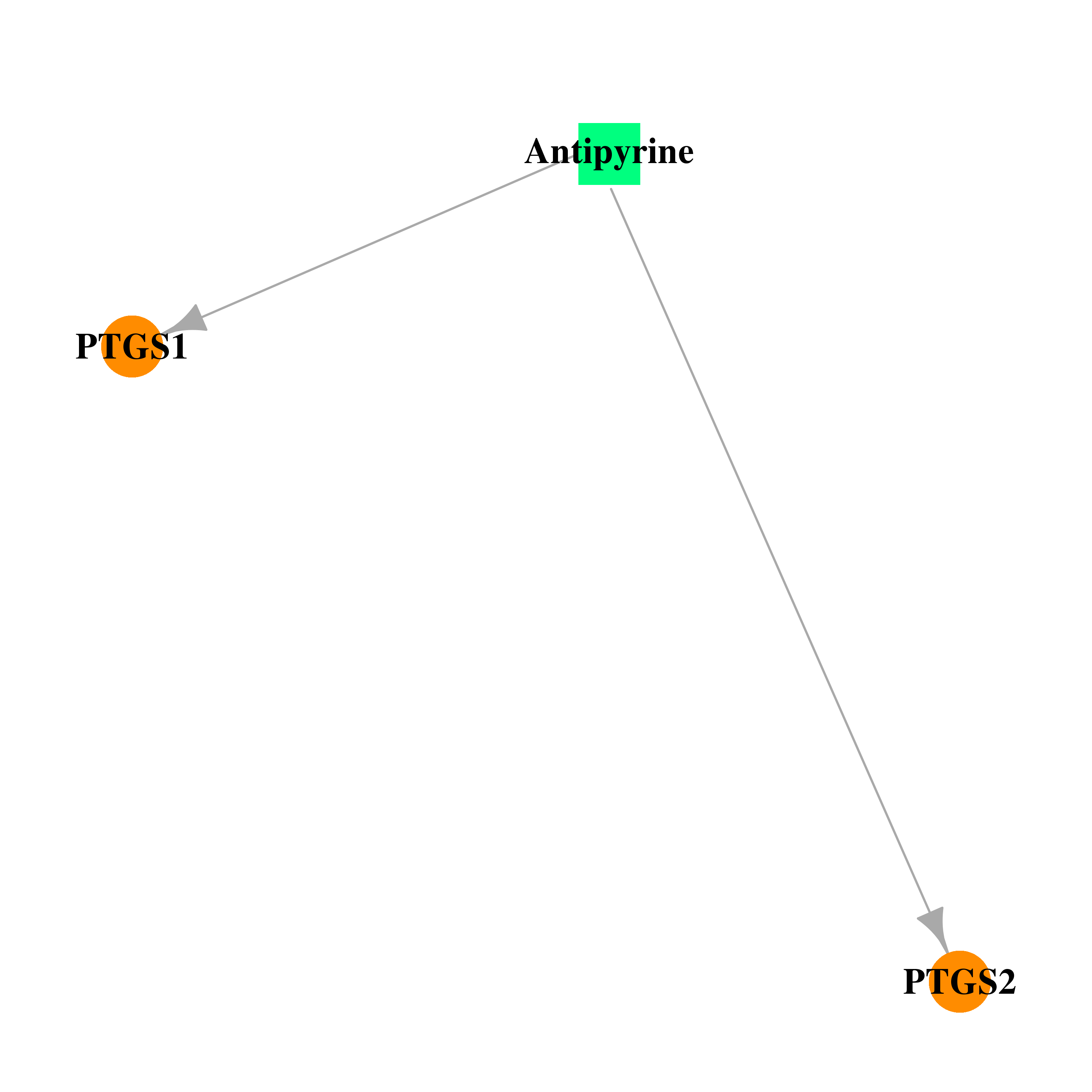 | 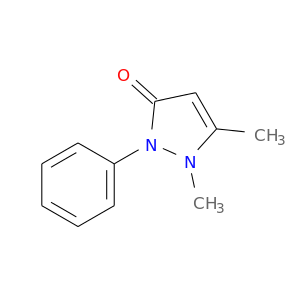 |
| DB01600 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Tiaprofenic acid | 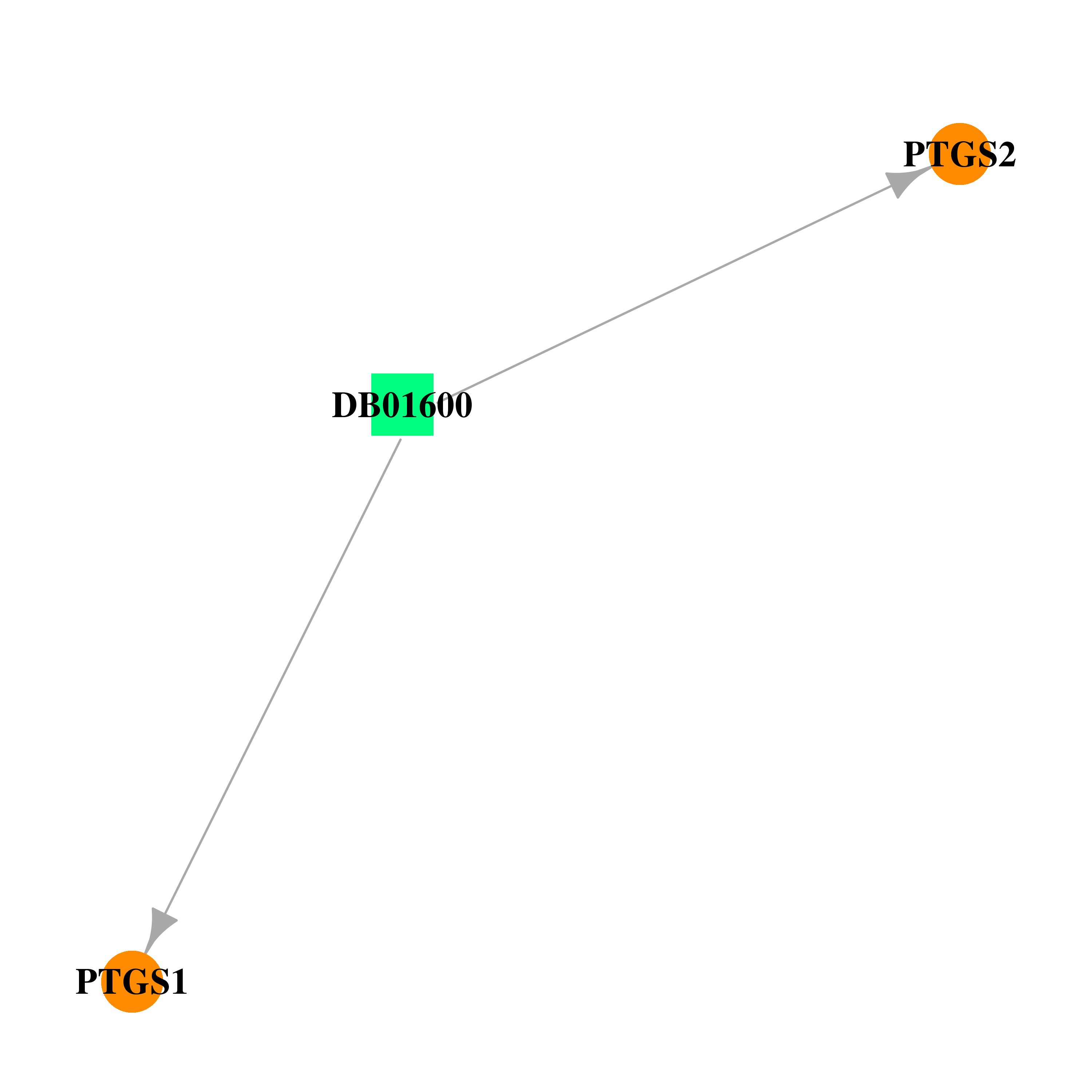 | 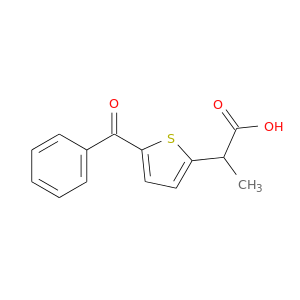 |
| DB01837 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | O-Acetylserine |  |  |
| DB01892 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | nutraceutical | Hyperforin |  |  |
| DB02047 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | 2-(1,1'-Biphenyl-4-Yl)Propanoic Acid |  |  |
| DB02110 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | Protoporphyrin Ix Containing Co |  |  |
| DB02198 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | 2-Bromoacetyl Group |  |  |
| DB02266 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | Flufenamic Acid |  |  |
| DB02379 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | Beta-D-Glucose |  |  |
| DB02687 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | Beta-D-Mannose |  |  |
| DB02709 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental; investigational | Resveratrol |  |  |
| DB02773 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | (3-Chloro-4-Propoxy-Phenyl)-Acetic Acid |  |  |
| DB02944 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | Alpha-D-Mannose |  |  |
| DB03014 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | Heme | 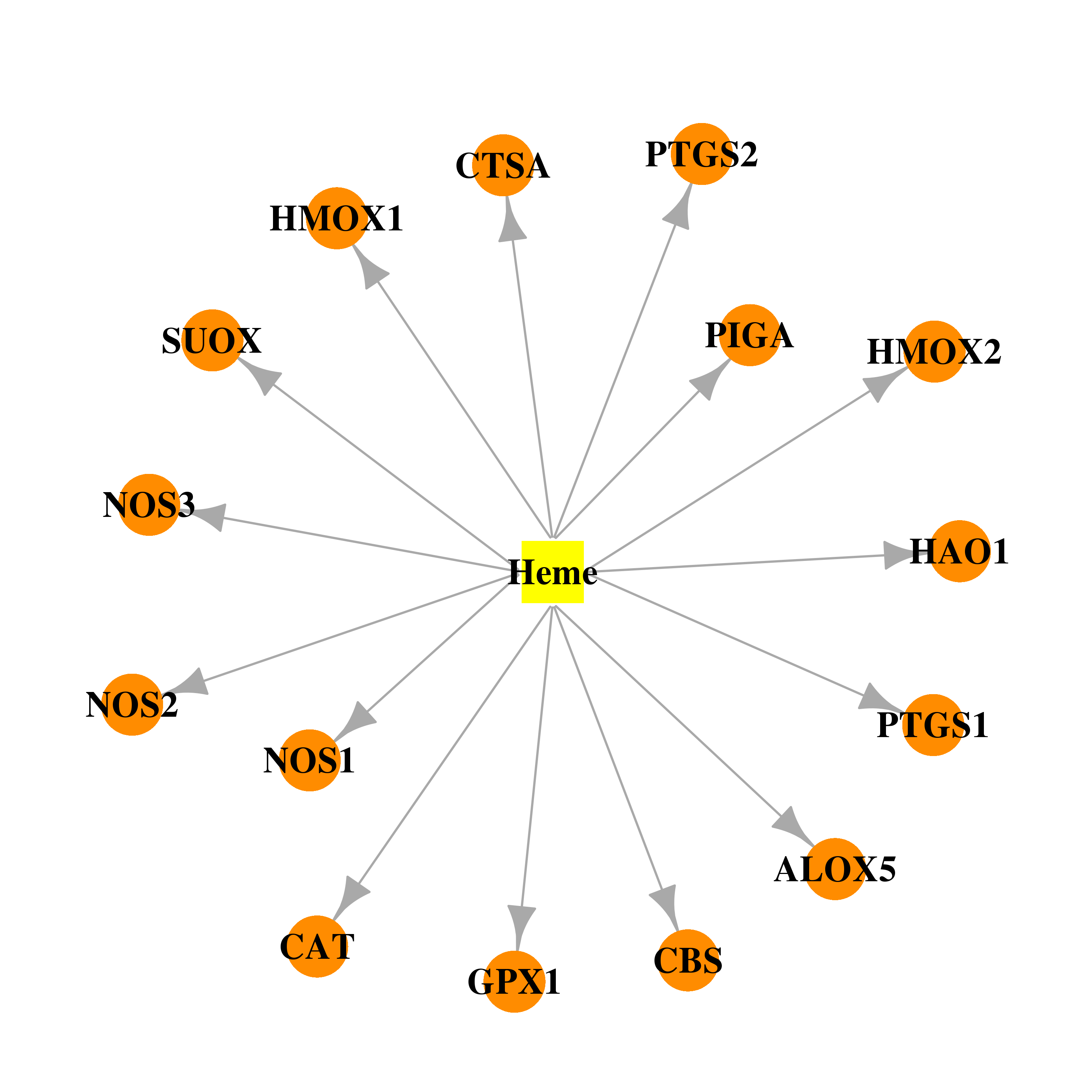 |  |
| DB03667 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | Acetic Acid Salicyloyl-Amino-Ester |  |  |
| DB03752 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | P-(2'-Iodo-5'-Thenoyl)Hydrotropic Acid |  |  |
| DB03753 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | Flurbiprofen Methyl Ester |  |  |
| DB03783 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental; withdrawn | Phenacetin |  |  |
| DB04552 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Niflumic Acid |  |  |
| DB04557 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | Arachidonic Acid |  |  |
| DB04817 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | withdrawn | Dipyrone |  |  |
| DB06725 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Lornoxicam |  | 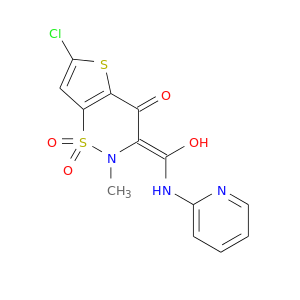 |
| DB06802 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Nepafenac |  |  |
| DB07981 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | 2-[1-(4-CHLOROBENZOYL)-5-METHOXY-2-METHYL-1H-INDOL-3-YL]-N-[(1R)-1-(HYDROXYMETHYL)PROPYL]ACETAMIDE |  |  |
| DB07983 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | 1-(4-IODOBENZOYL)-5-METHOXY-2-METHYL INDOLE-3-ACETIC ACID |  |  |
| DB07984 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | experimental | 2-[1-(4-CHLOROBENZOYL)-5-METHOXY-2-METHYL-1H-INDOL-3-YL]-N-[(1S)-1-(HYDROXYMETHYL)PROPYL]ACETAMIDE |  |  |
| DB00532 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Mephenytoin | 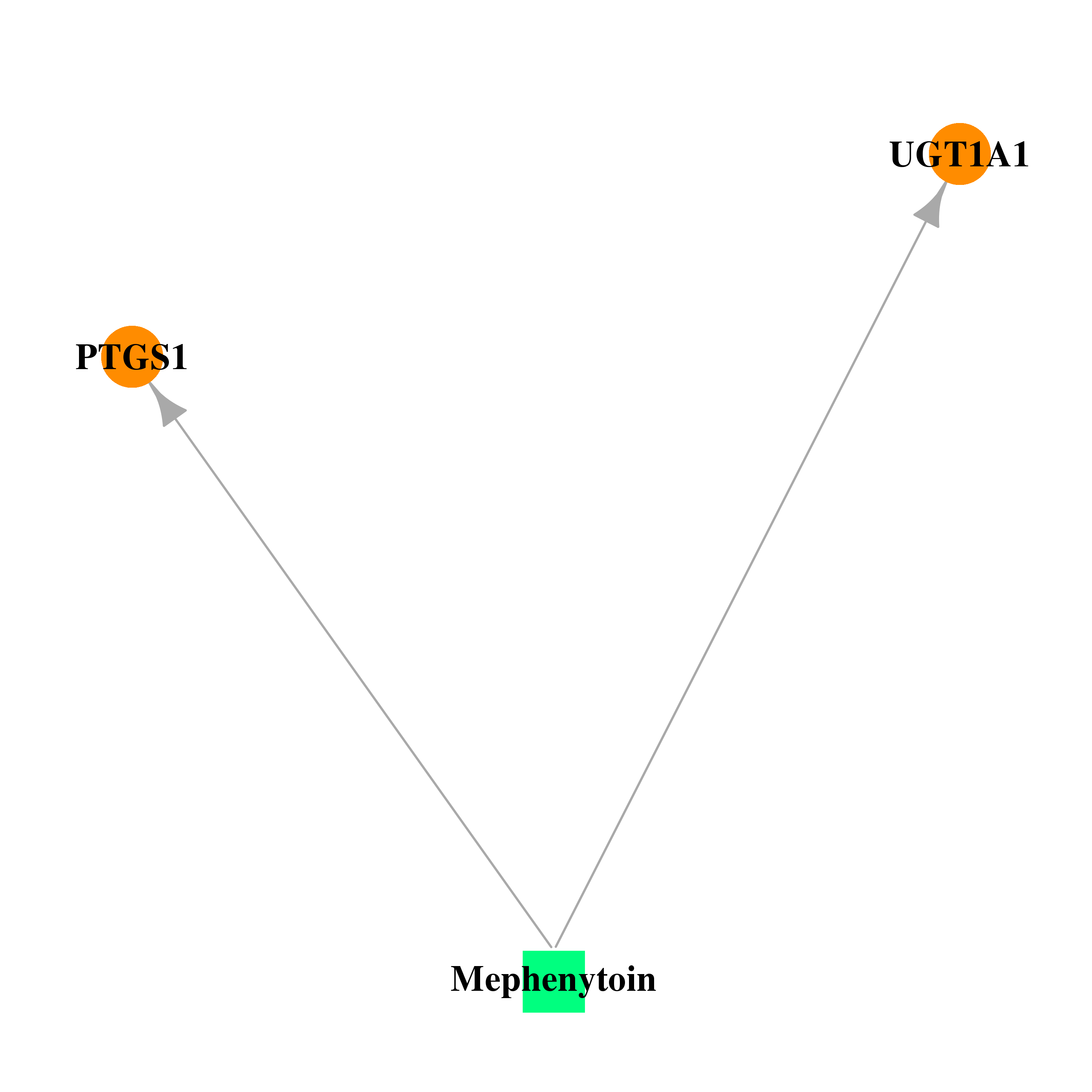 |  |
| DB01174 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Phenobarbital | 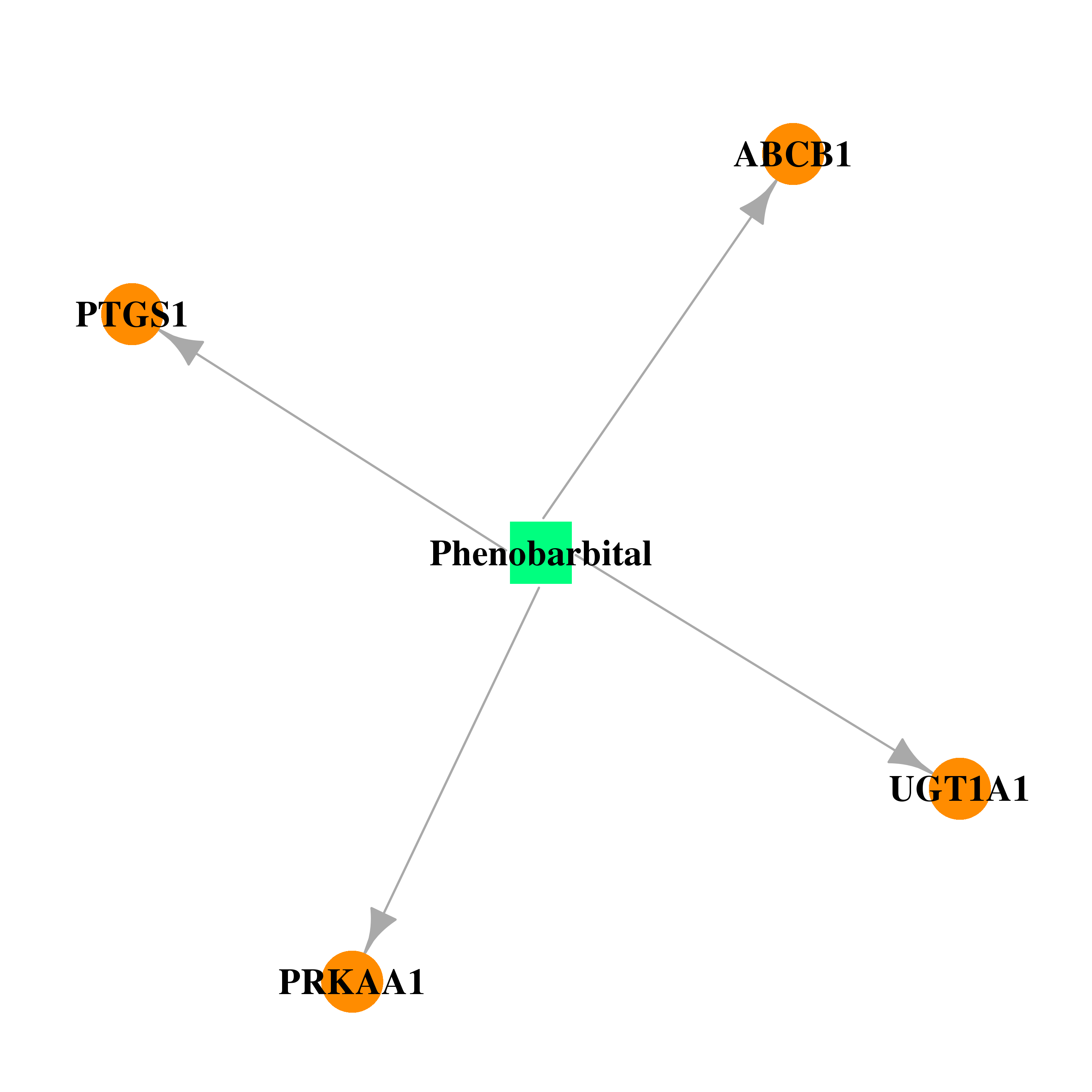 |  |
| DB00252 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Phenytoin | 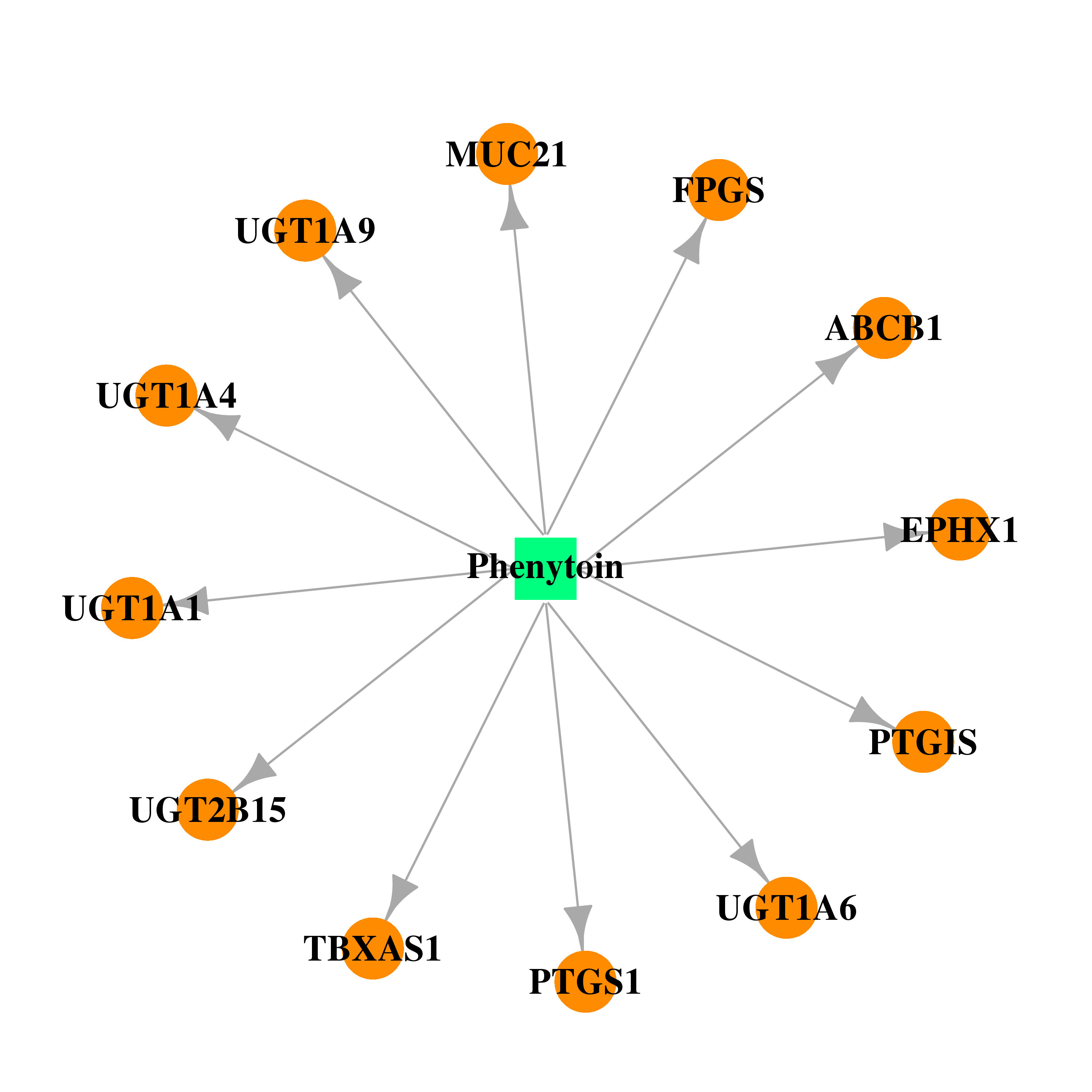 |  |
| DB00347 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Trimethadione | 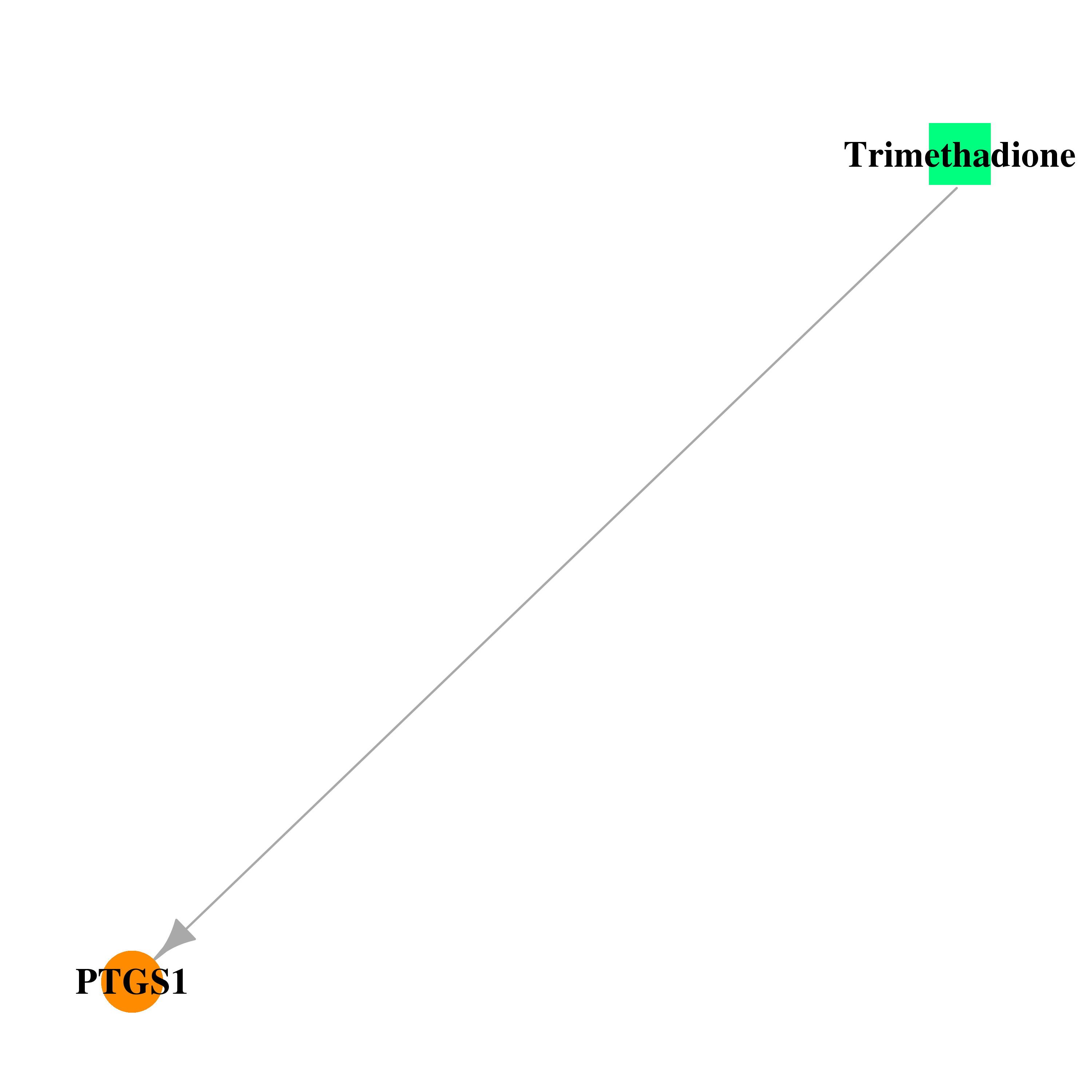 |  |
| DB00917 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Dinoprostone | 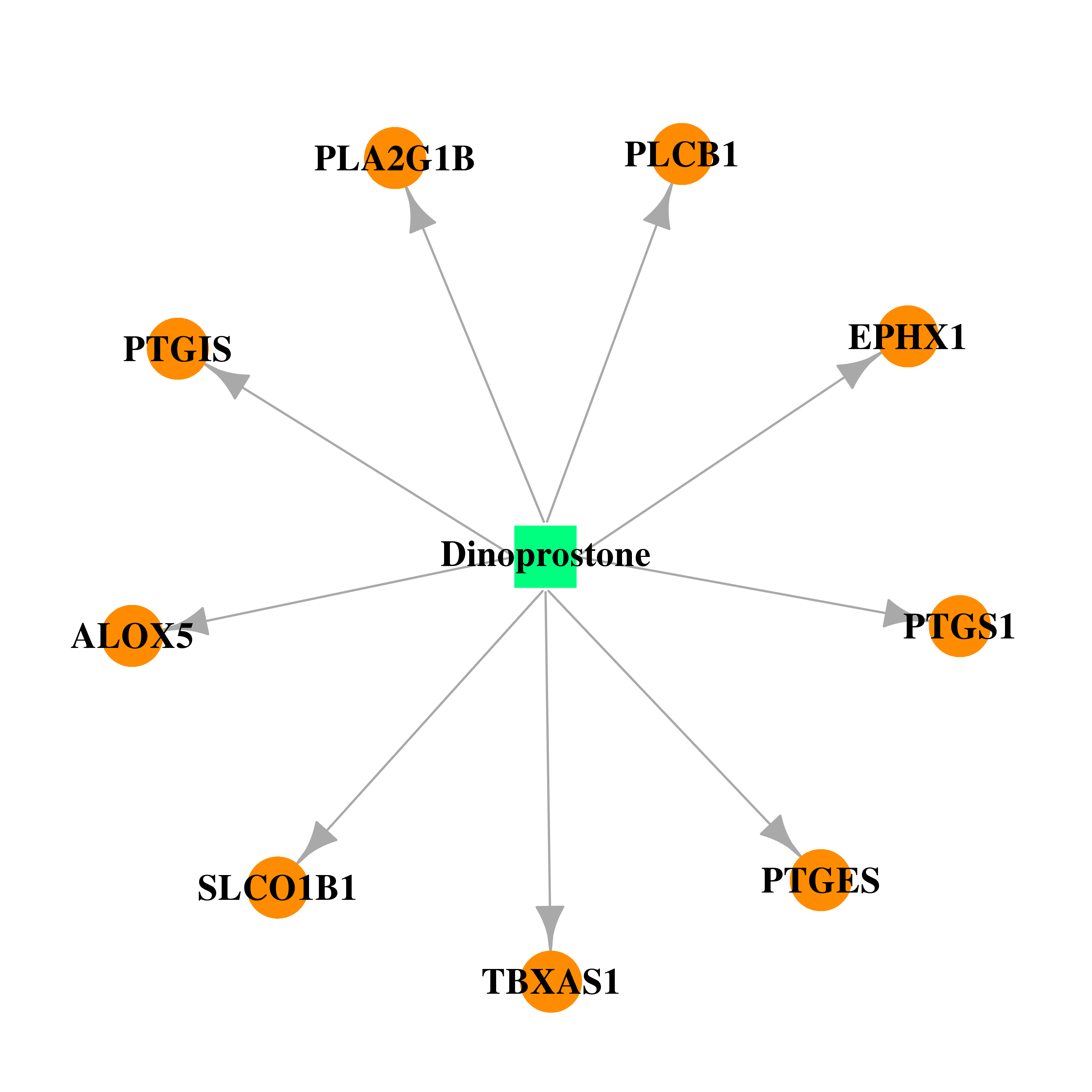 |  |
| DB01240 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved | Epoprostenol | 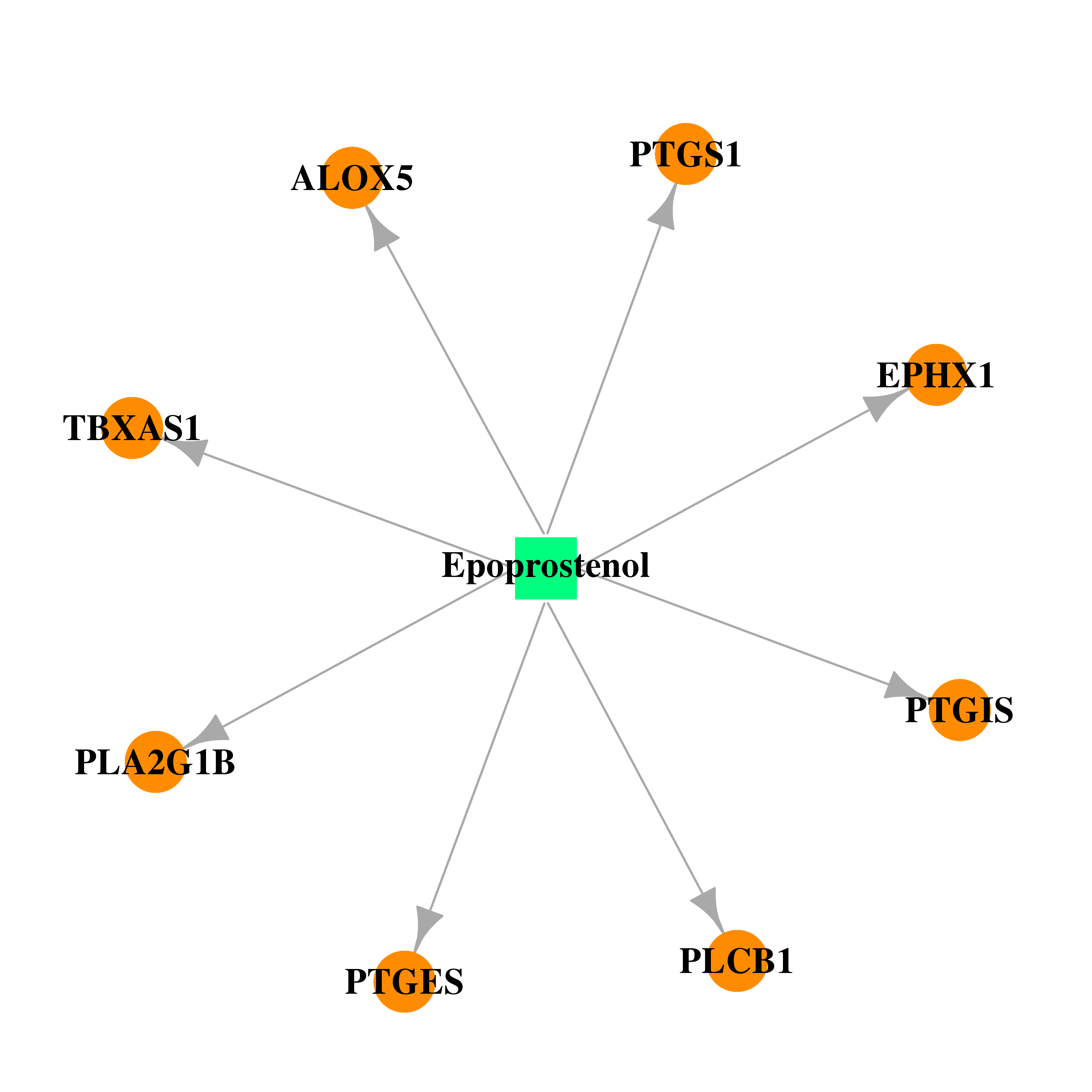 | 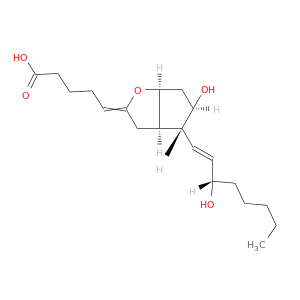 |
| DB00482 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | approved; investigational | Celecoxib | 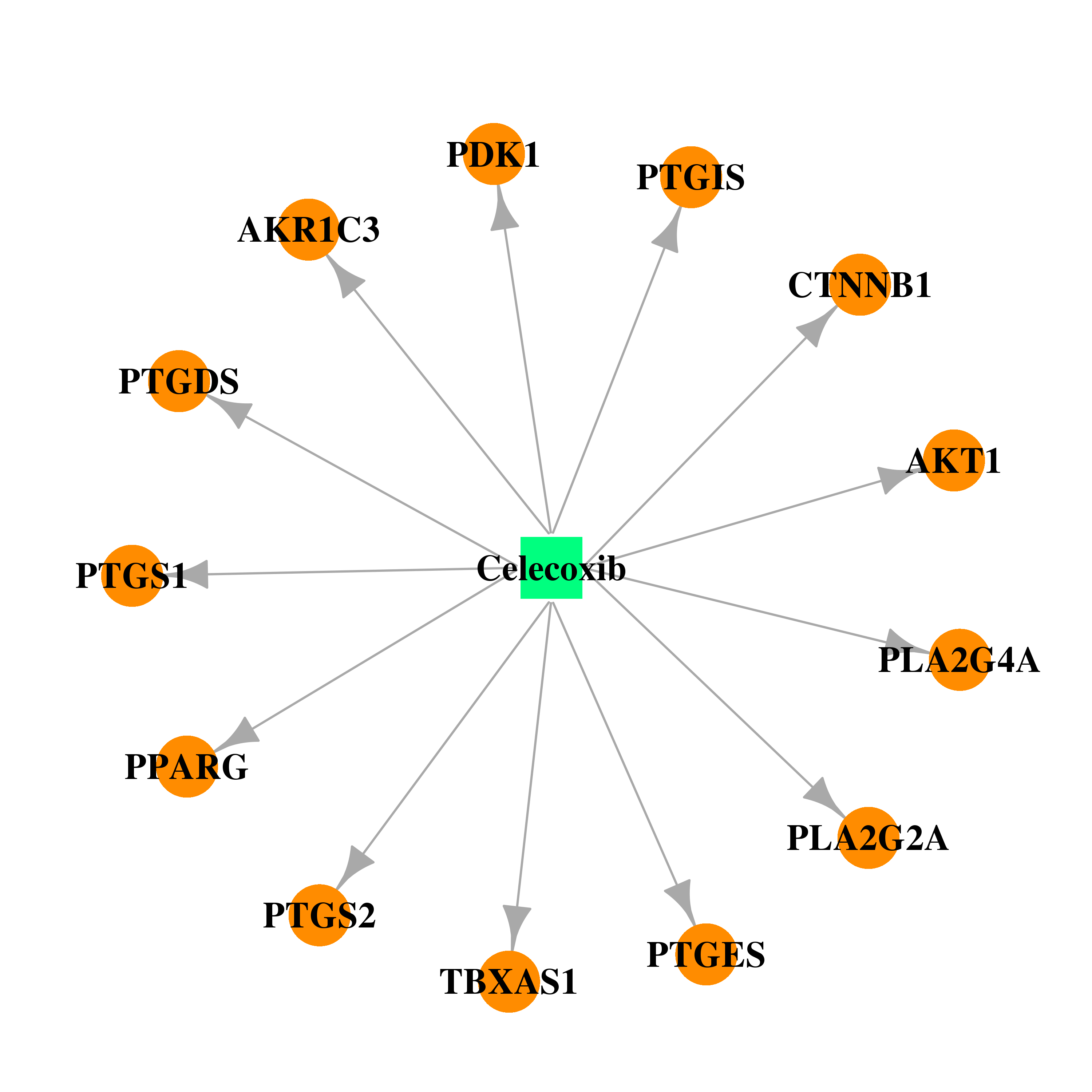 |  |