|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for CA2 |
| Basic gene info. | Gene symbol | CA2 |
| Gene name | carbonic anhydrase II | |
| Synonyms | CA-II|CAC|CAII|Car2|HEL-76 | |
| Cytomap | UCSC genome browser: 8q22 | |
| Genomic location | chr8 :86376130-86393721 | |
| Type of gene | protein-coding | |
| RefGenes | NM_000067.2, NM_001293675.1, | |
| Ensembl id | ENSG00000104267 | |
| Description | carbonate dehydratase IIcarbonic anhydrase 2carbonic anhydrase Bcarbonic anhydrase Ccarbonic dehydrataseepididymis luminal protein 76 | |
| Modification date | 20141221 | |
| dbXrefs | MIM : 611492 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000104267 | ||
| HPRD : 02023 | ||
| Vega : OTTHUMG00000164944 | ||
| Protein | UniProt: P00918 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_CA2 | |
| BioGPS: 760 | ||
| Gene Expression Atlas: ENSG00000104267 | ||
| The Human Protein Atlas: ENSG00000104267 | ||
| Pathway | NCI Pathway Interaction Database: CA2 | |
| KEGG: CA2 | ||
| REACTOME: CA2 | ||
| ConsensusPathDB | ||
| Pathway Commons: CA2 | ||
| Metabolism | MetaCyc: CA2 | |
| HUMANCyc: CA2 | ||
| Regulation | Ensembl's Regulation: ENSG00000104267 | |
| miRBase: chr8 :86,376,130-86,393,721 | ||
| TargetScan: NM_000067 | ||
| cisRED: ENSG00000104267 | ||
| Context | iHOP: CA2 | |
| cancer metabolism search in PubMed: CA2 | ||
| UCL Cancer Institute: CA2 | ||
| Assigned class in ccmGDB | A - This gene has a literature evidence and it belongs to cancer gene. | |
| References showing role of CA2 in cancer cell metabolism | 1. Sheng W, Dong M, Zhou J, Li X, Dong Q (2013) Down regulation of CAII is associated with tumor differentiation and poor prognosis in patients with pancreatic cancer. J Surg Oncol 107: 536-543. doi: 10.1002/jso.23282. go to article 2. Zhou R, Huang W, Yao Y, Wang Y, Li Z, et al. (2013) CA II, a potential biomarker by proteomic analysis, exerts significant inhibitory effect on the growth of colorectal cancer cells. Int J Oncol 43: 611-621. doi: 10.3892/ijo.2013.1972. go to article | |
| Top |
| Phenotypic Information for CA2(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: CA2 |
| Familial Cancer Database: CA2 | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Oncogene 1 | Significant driver gene in | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| KEGG_NITROGEN_METABOLISM | |
| OMIM | 259730; phenotype. 611492; gene. |
| Orphanet | 2785; Osteopetrosis with renal tubular acidosis. |
| Disease | KEGG Disease: CA2 |
| MedGen: CA2 (Human Medical Genetics with Condition) | |
| ClinVar: CA2 | |
| Phenotype | MGI: CA2 (International Mouse Phenotyping Consortium) |
| PhenomicDB: CA2 | |
| Mutations for CA2 |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| - Statistics for Tissue and Mutation type | Top |
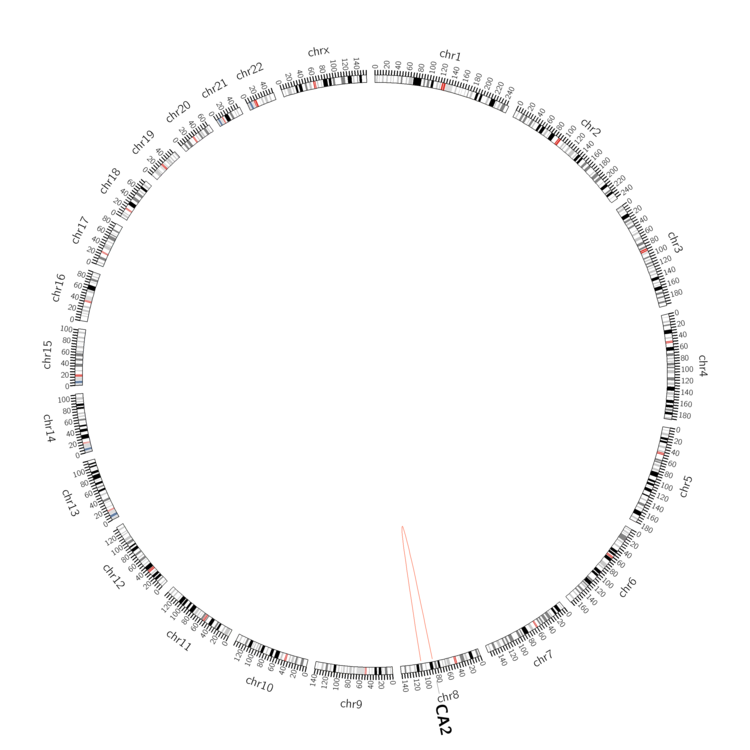 |
| - For Inter-chromosomal Variations |
| There's no inter-chromosomal structural variation. |
| - For Intra-chromosomal Variations |
| * Intra-chromosomal variantions includes 'intrachromosomal amplicon to amplicon', 'intrachromosomal amplicon to non-amplified dna', 'intrachromosomal deletion', 'intrachromosomal fold-back inversion', 'intrachromosomal inversion', 'intrachromosomal tandem duplication', 'Intrachromosomal unknown type', 'intrachromosomal with inverted orientation', 'intrachromosomal with non-inverted orientation'. |
 |
| Sample | Symbol_a | Chr_a | Start_a | End_a | Symbol_b | Chr_b | Start_b | End_b |
| ovary | CA2 | chr8 | 86385648 | 86385668 | chr8 | 107841578 | 107841598 | |
| ovary | CA2 | chr8 | 86385649 | 86385669 | chr8 | 107841772 | 107841792 | |
| ovary | CA2 | chr8 | 86385649 | 86385669 | chr8 | 107841776 | 107841796 |
| cf) Tissue number; Tissue name (1;Breast, 2;Central_nervous_system, 3;Haematopoietic_and_lymphoid_tissue, 4;Large_intestine, 5;Liver, 6;Lung, 7;Ovary, 8;Pancreas, 9;Prostate, 10;Skin, 11;Soft_tissue, 12;Upper_aerodigestive_tract) |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows CA2 related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| BI042447 | MSC | 16 | 214 | 8 | 72754213 | 72754411 | CA2 | 212 | 369 | 8 | 86385949 | 86386618 | |
| Top |
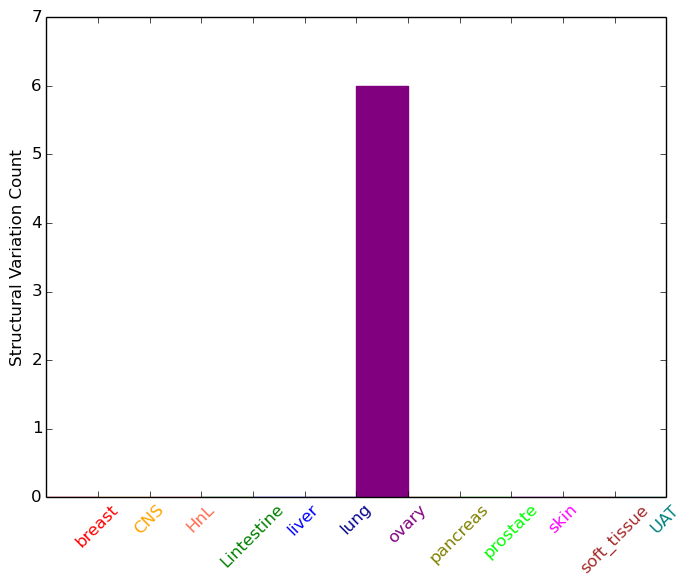
| Mutation type/ Tissue ID | brca | cns | cerv | endome | haematopo | kidn | Lintest | liver | lung | ns | ovary | pancre | prost | skin | stoma | thyro | urina | |||
| Total # sample | 1 | 1 | 1 | |||||||||||||||||
| GAIN (# sample) | 1 | 1 | 1 | |||||||||||||||||
| LOSS (# sample) |
| cf) Tissue ID; Tissue type (1; Breast, 2; Central_nervous_system, 3; Cervix, 4; Endometrium, 5; Haematopoietic_and_lymphoid_tissue, 6; Kidney, 7; Large_intestine, 8; Liver, 9; Lung, 10; NS, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary_tract) |
| Top |
|
 |
| Top |
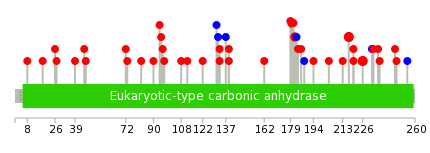
| Stat. for Non-Synonymous SNVs (# total SNVs=23) | (# total SNVs=1) |
 |  |
(# total SNVs=1) | (# total SNVs=0) |
 |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr8:86389490-86389490 | p.V217I | 3 |
| chr8:86392911-86392911 | p.R226C | 3 |
| chr8:86389380-86389380 | p.P180H | 2 |
| chr8:86389385-86389385 | p.G182R | 2 |
| chr8:86392932-86392932 | p.E233K | 2 |
| chr8:86392979-86392979 | p.Q248H | 1 |
| chr8:86386011-86386011 | p.T108S | 1 |
| chr8:86388066-86388066 | p.V162L | 1 |
| chr8:86389478-86389478 | p.E213K | 1 |
| chr8:86377599-86377599 | p.K45E | 1 |
| Top |
|
 |
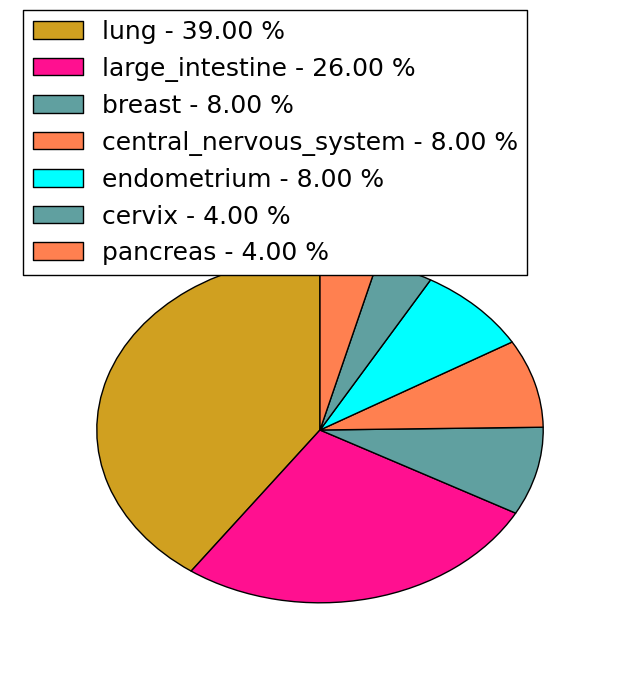
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 1 | 1 | 3 | 3 | 3 | 1 | 11 | 3 | 2 | |||||||||||
| # mutation | 1 | 1 | 3 | 3 | 3 | 1 | 13 | 3 | 2 | |||||||||||
| nonsynonymous SNV | 1 | 1 | 3 | 1 | 3 | 1 | 9 | 2 | 2 | |||||||||||
| synonymous SNV | 2 | 4 | 1 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr8:86389385 | p.G81R,CA2 | 2 |
| chr8:86386612 | p.V105V,CA2 | 1 |
| chr8:86392938 | p.H96Q | 1 |
| chr8:86385963 | p.W107C,CA2 | 1 |
| chr8:86386616 | p.W97L | 1 |
| chr8:86392975 | p.R125C,CA2 | 1 |
| chr8:86385977 | p.K112N | 1 |
| chr8:86389382 | p.G131G,CA2 | 1 |
| chr8:86385979 | p.H21Y,CA2 | 1 |
| chr8:86386025 | p.E134K,CA2 | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
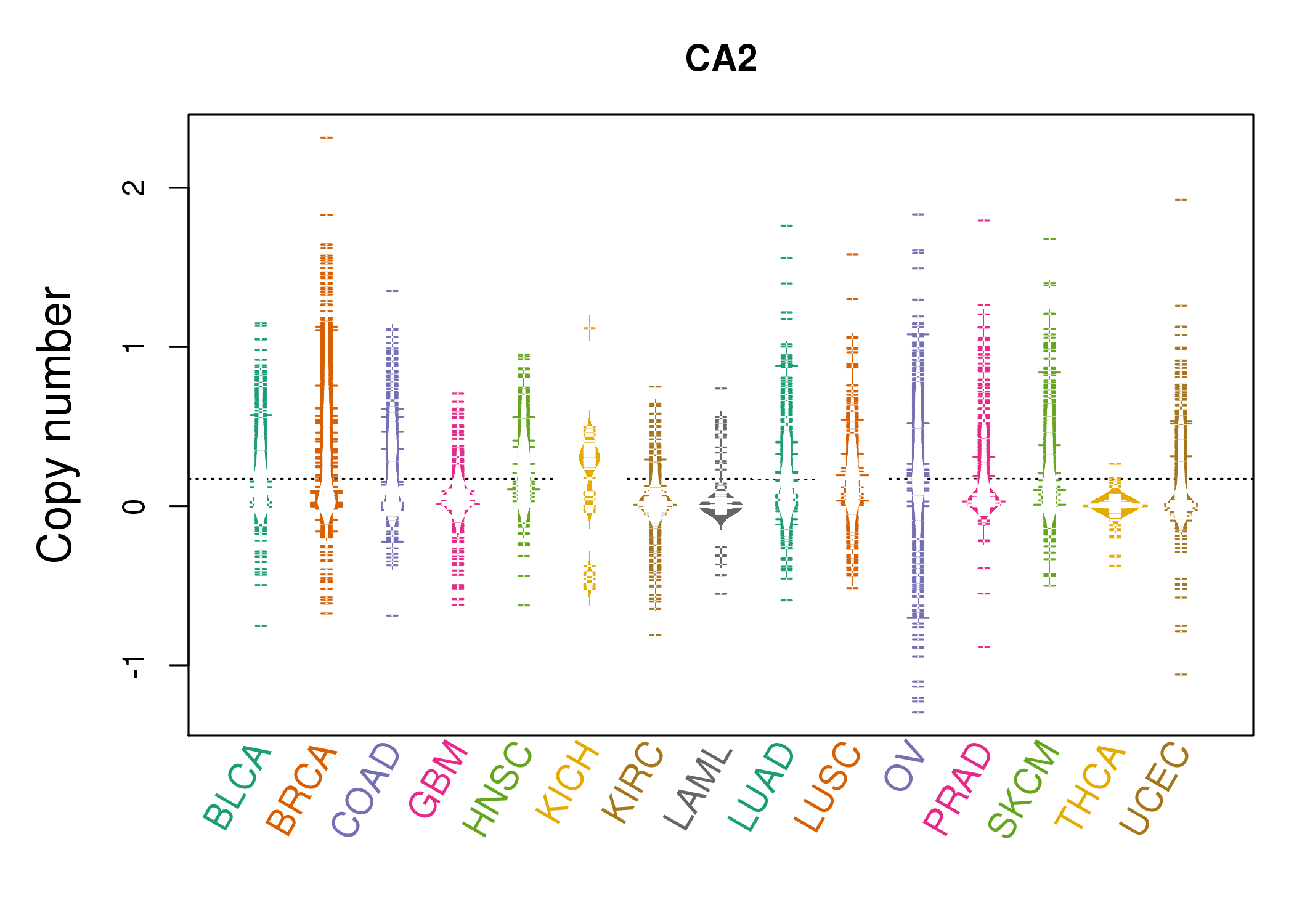 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
| Gene Expression for CA2 |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
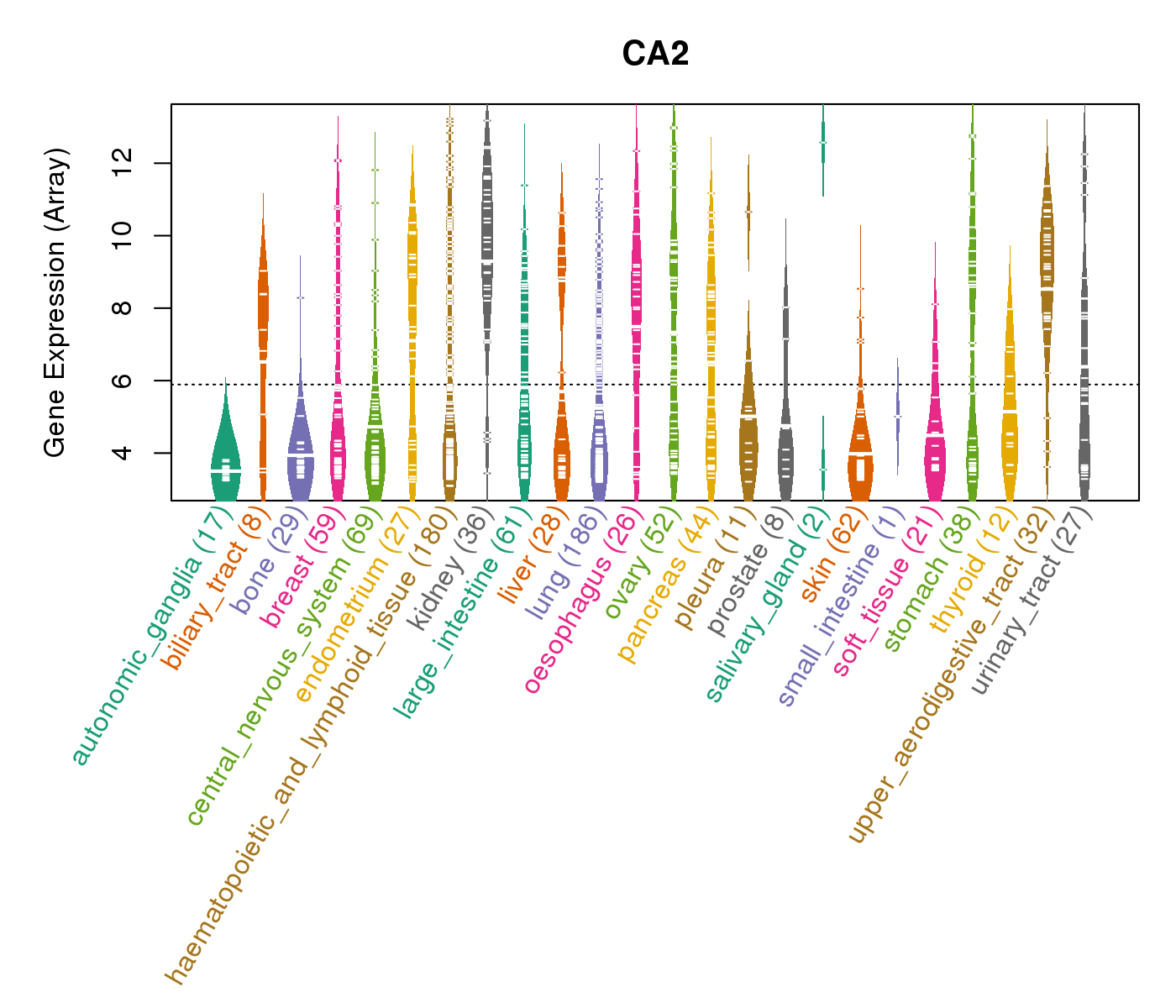 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
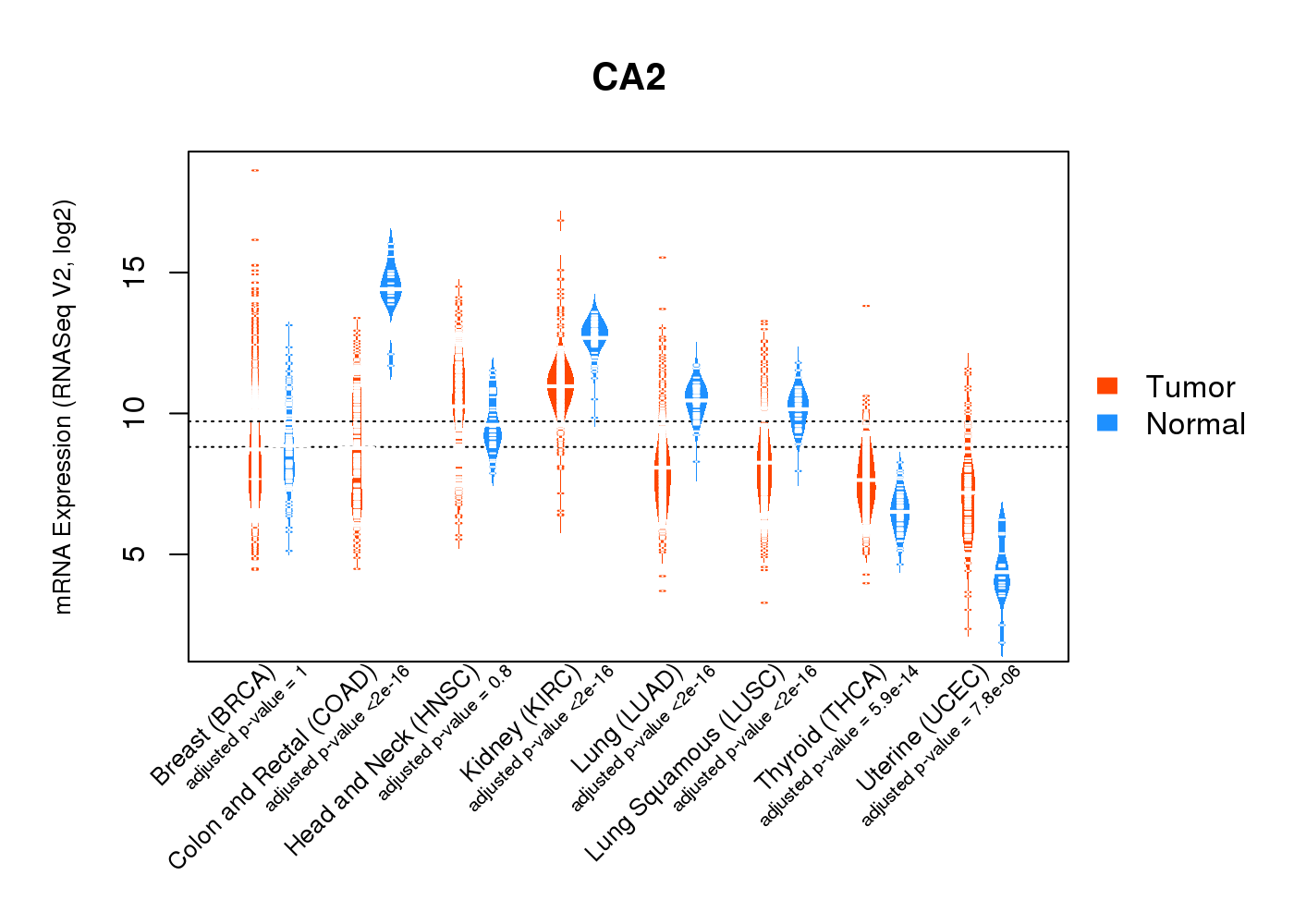 |
| Top |
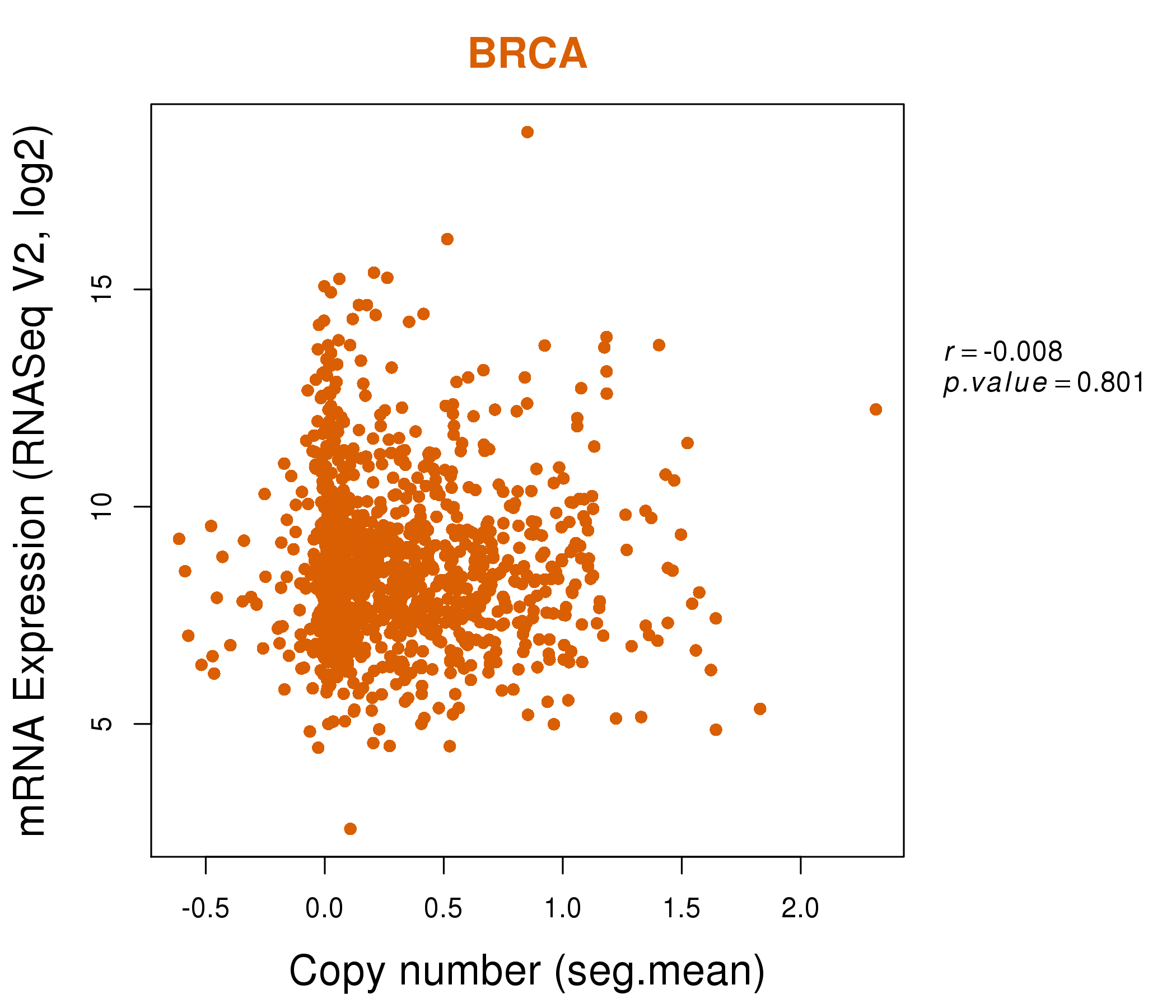
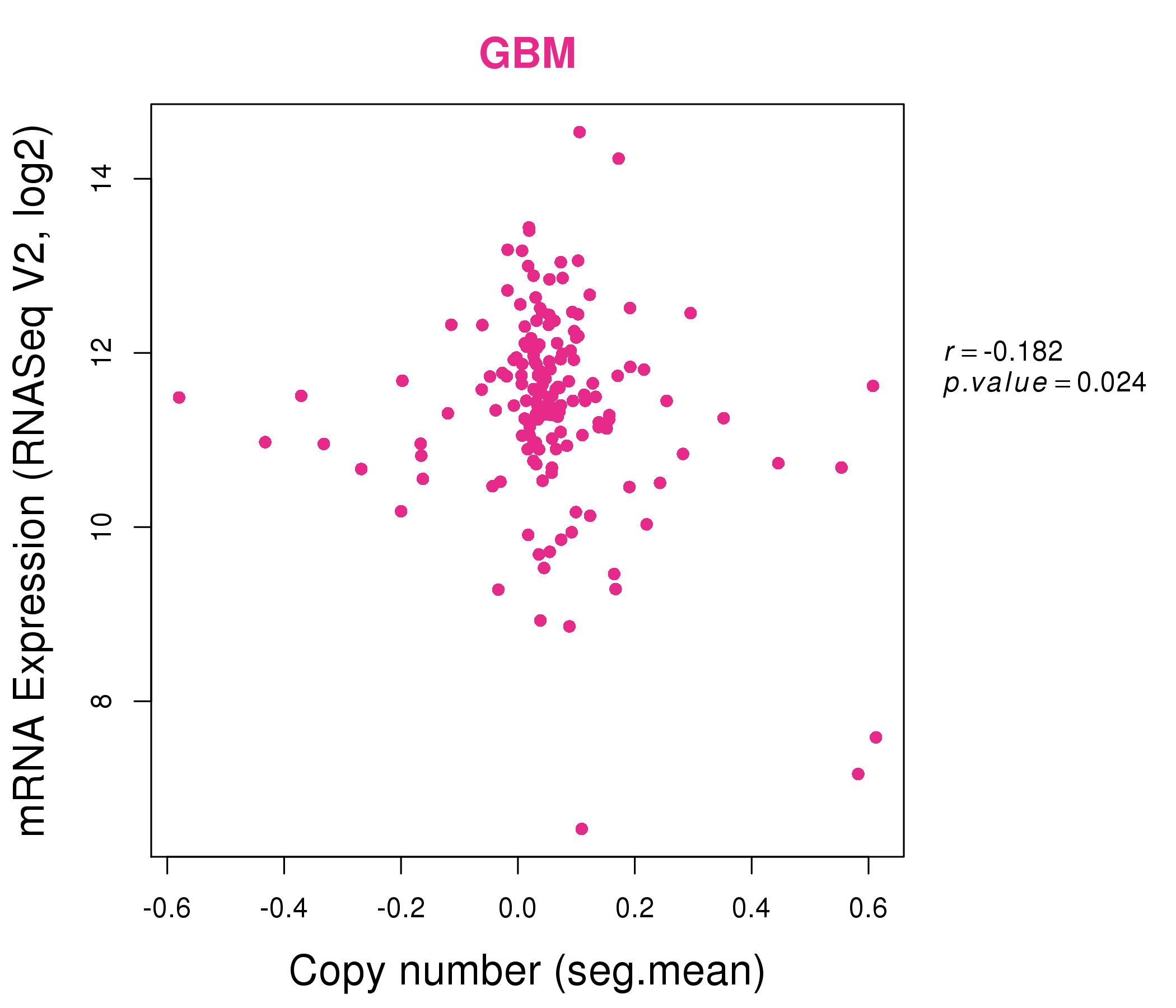
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
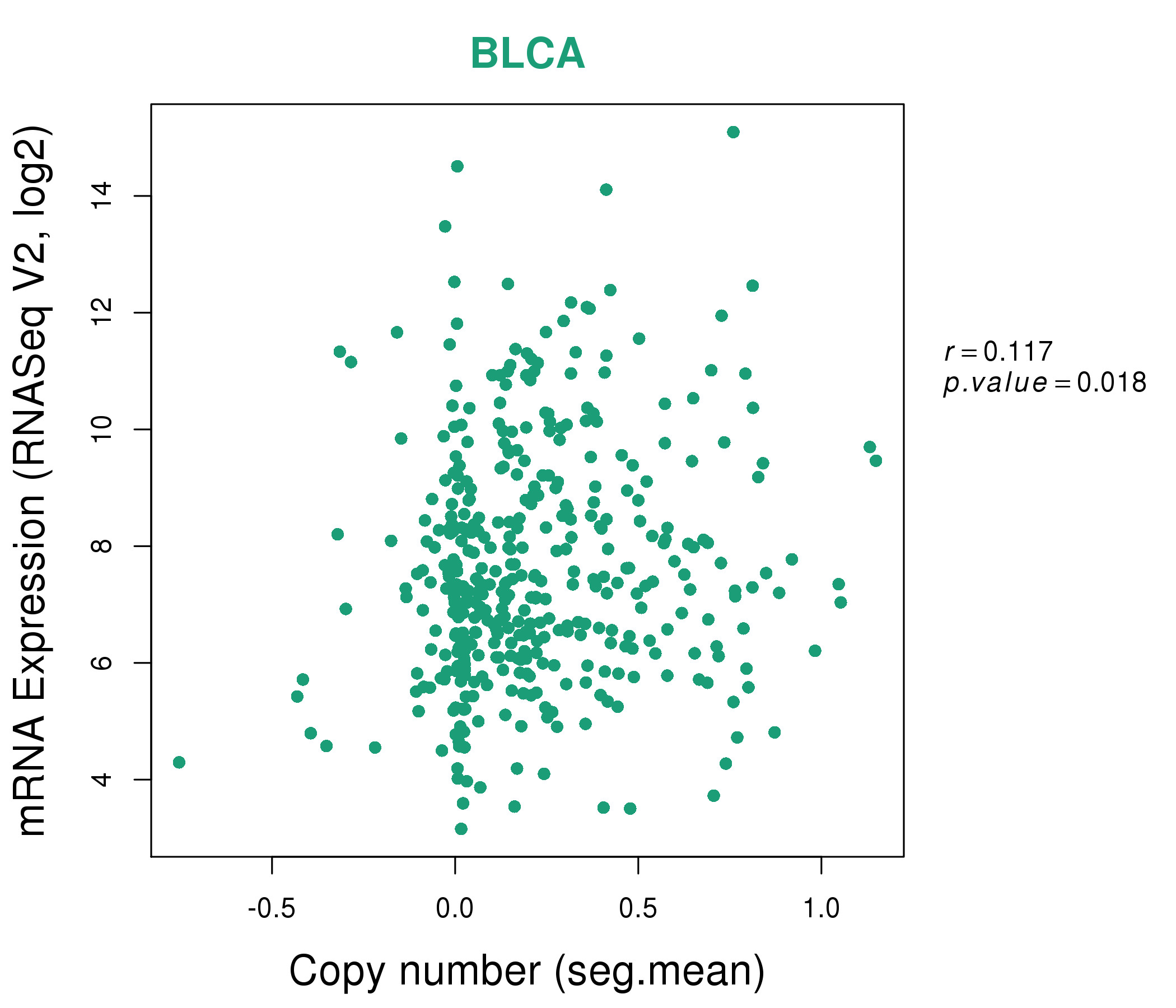 |
|
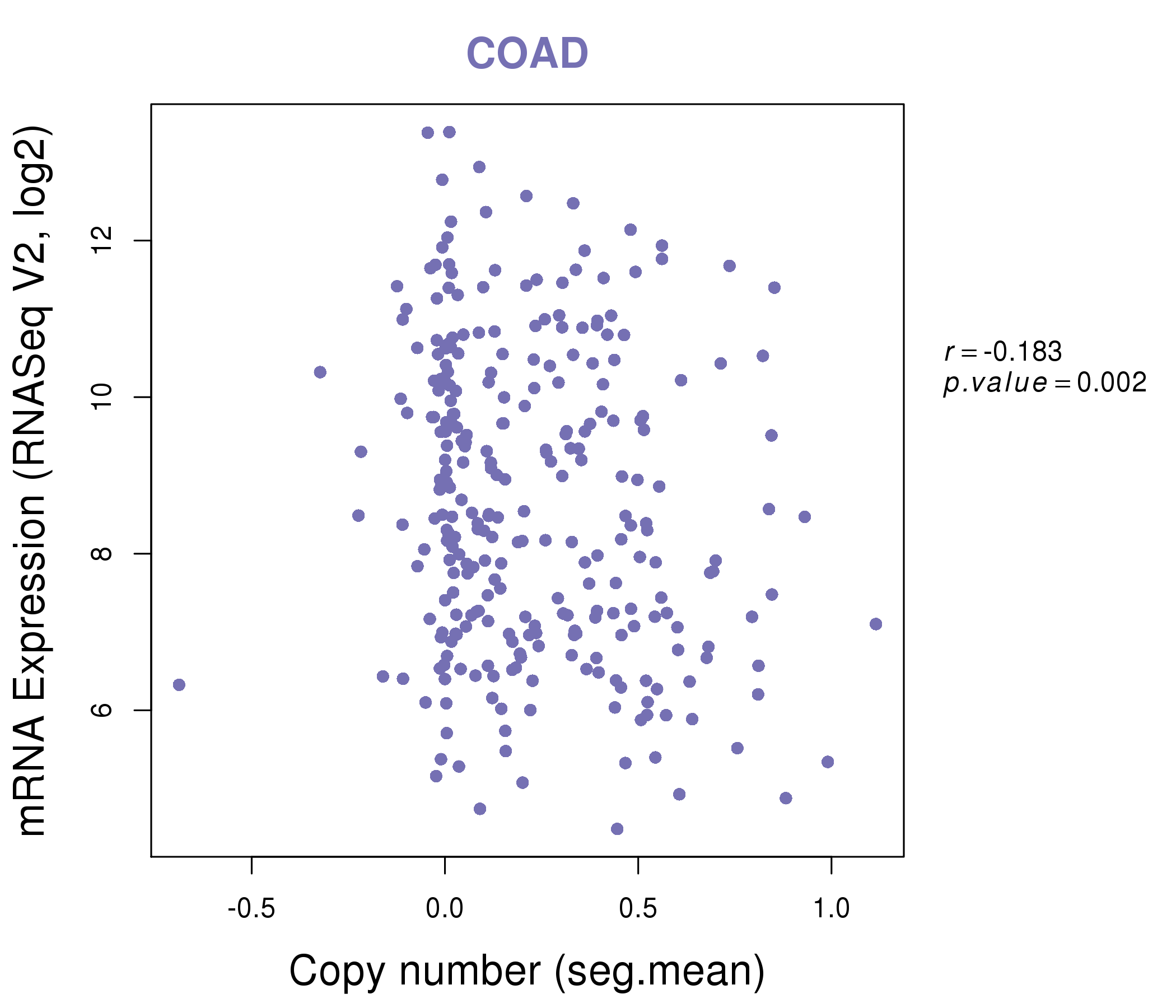 |
|
| Top |
| Gene-Gene Network Information |
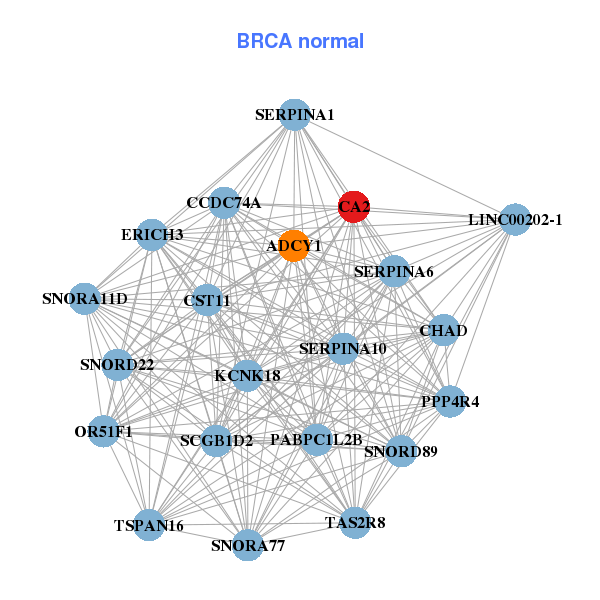
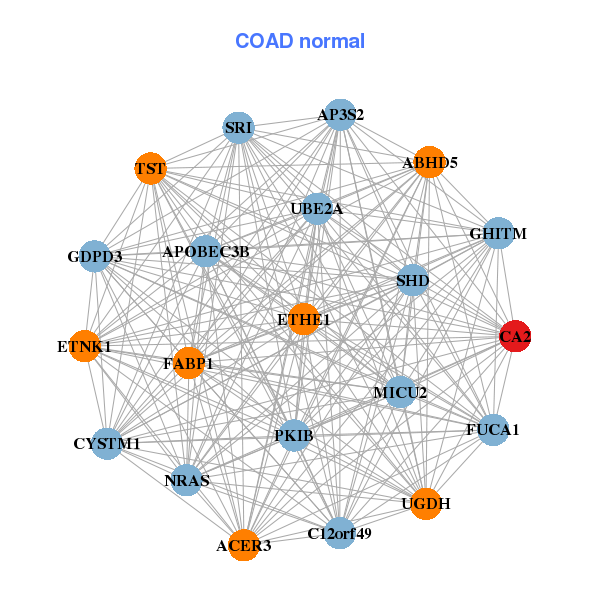
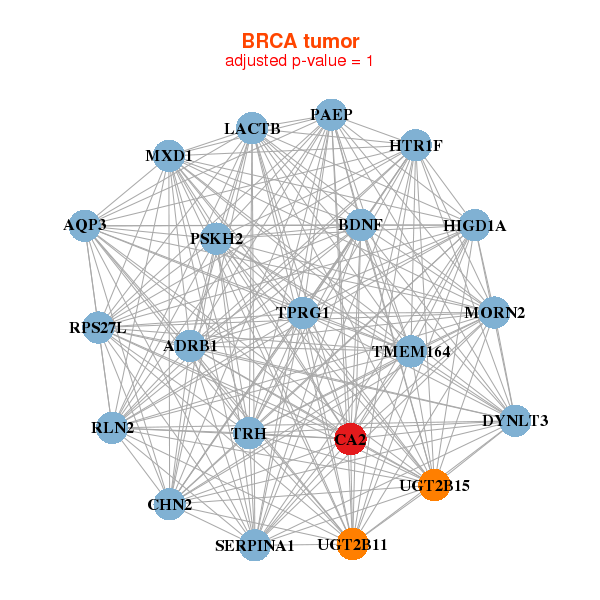
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
 |
| ||||
| ADRB1,AQP3,BDNF,CA2,CHN2,DYNLT3,HIGD1A, HTR1F,LACTB,MORN2,MXD1,PAEP,PSKH2,RLN2, RPS27L,SERPINA1,TMEM164,TPRG1,TRH,UGT2B11,UGT2B15 | ADCY1,ERICH3,CA2,CCDC74A,CHAD,CST11,KCNK18, LINC00202-1,OR51F1,PABPC1L2B,PPP4R4,SCGB1D2,SERPINA10,SERPINA1, SERPINA6,SNORA11D,SNORA77,SNORD22,SNORD89,TAS2R8,TSPAN16 | ||||
 |
| ||||
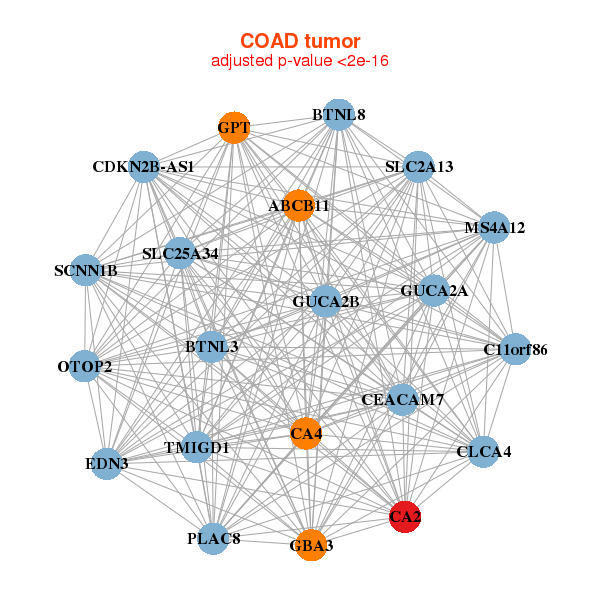
| ABCB11,BTNL3,BTNL8,C11orf86,CA2,CA4,CDKN2B-AS1, CEACAM7,CLCA4,EDN3,GBA3,GPT,GUCA2A,GUCA2B, MS4A12,OTOP2,PLAC8,SCNN1B,SLC25A34,SLC2A13,TMIGD1 | ABHD5,ACER3,AP3S2,APOBEC3B,C12orf49,CYSTM1,CA2, MICU2,ETHE1,ETNK1,FABP1,FUCA1,GDPD3,GHITM, NRAS,PKIB,SHD,SRI,TST,UBE2A,UGDH |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for CA2 |
| DB Category | DB Name | DB's ID and Url link |
| Chemistry | BindingDB | P00918; -. |
| Chemistry | ChEMBL | CHEMBL205; -. |
| Organism-specific databases | PharmGKB | PA25989; -. |
| Organism-specific databases | CTD | 760; -. |
| * Gene Centered Interaction Network. |
 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB00232 | carbonic anhydrase II | approved | Methyclothiazide |  |  |
| DB00273 | carbonic anhydrase II | approved; investigational | Topiramate | 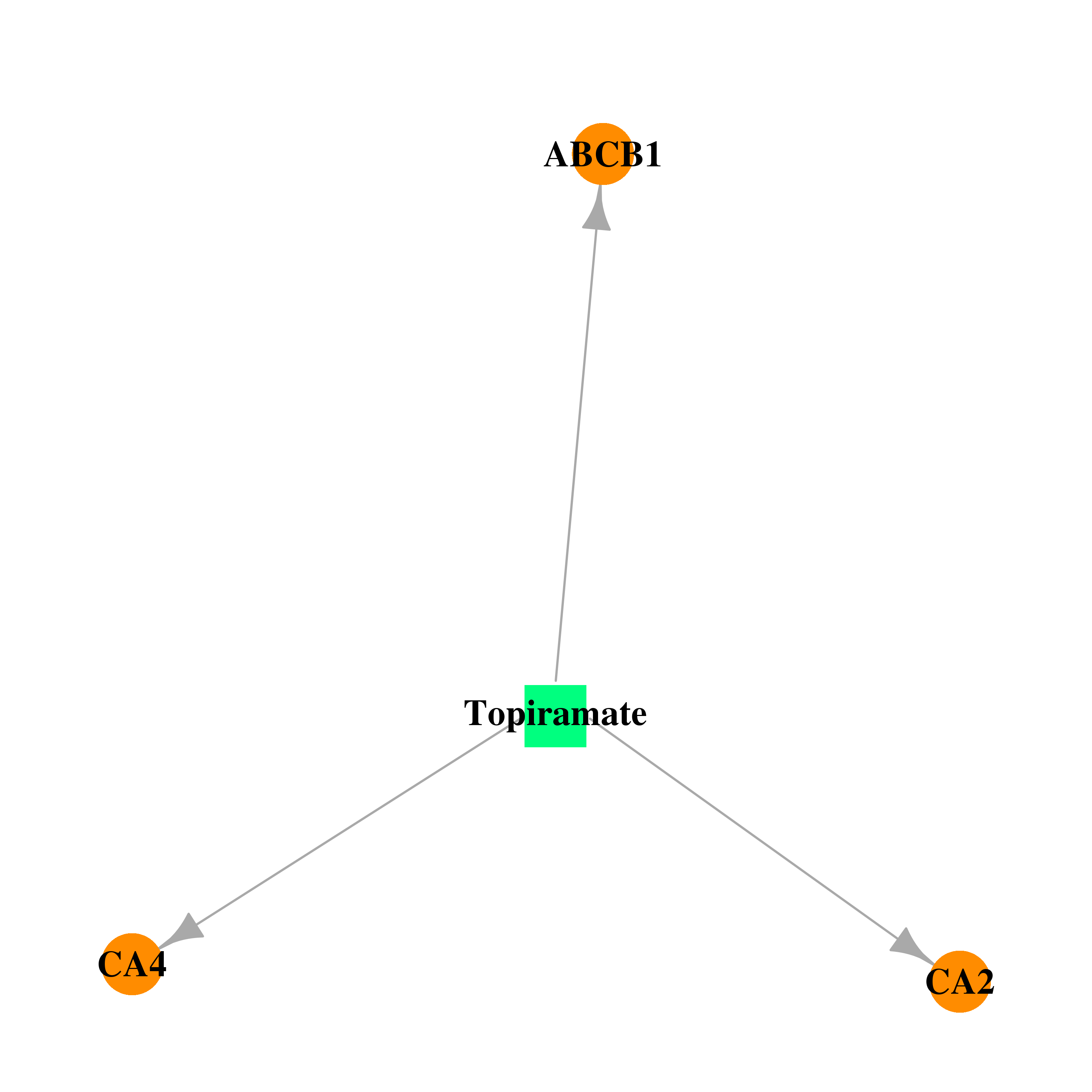 |  |
| DB00311 | carbonic anhydrase II | approved | Ethoxzolamide |  |  |
| DB00436 | carbonic anhydrase II | approved | Bendroflumethiazide |  |  |
| DB00562 | carbonic anhydrase II | approved | Benzthiazide |  |  |
| DB00606 | carbonic anhydrase II | approved | Cyclothiazide |  |  |
| DB00695 | carbonic anhydrase II | approved | Furosemide |  |  |
| DB00703 | carbonic anhydrase II | approved | Methazolamide |  |  |
| DB00774 | carbonic anhydrase II | approved | Hydroflumethiazide |  |  |
| DB00819 | carbonic anhydrase II | approved | Acetazolamide |  |  |
| DB00869 | carbonic anhydrase II | approved | Dorzolamide |  |  |
| DB00880 | carbonic anhydrase II | approved | Chlorothiazide |  |  |
| DB00909 | carbonic anhydrase II | approved; investigational | Zonisamide |  |  |
| DB00999 | carbonic anhydrase II | approved | Hydrochlorothiazide |  |  |
| DB01021 | carbonic anhydrase II | approved | Trichlormethiazide |  |  |
| DB01031 | carbonic anhydrase II | illicit; approved | Ethinamate |  |  |
| DB01119 | carbonic anhydrase II | approved | Diazoxide |  |  |
| DB01144 | carbonic anhydrase II | approved | Dichlorphenamide | 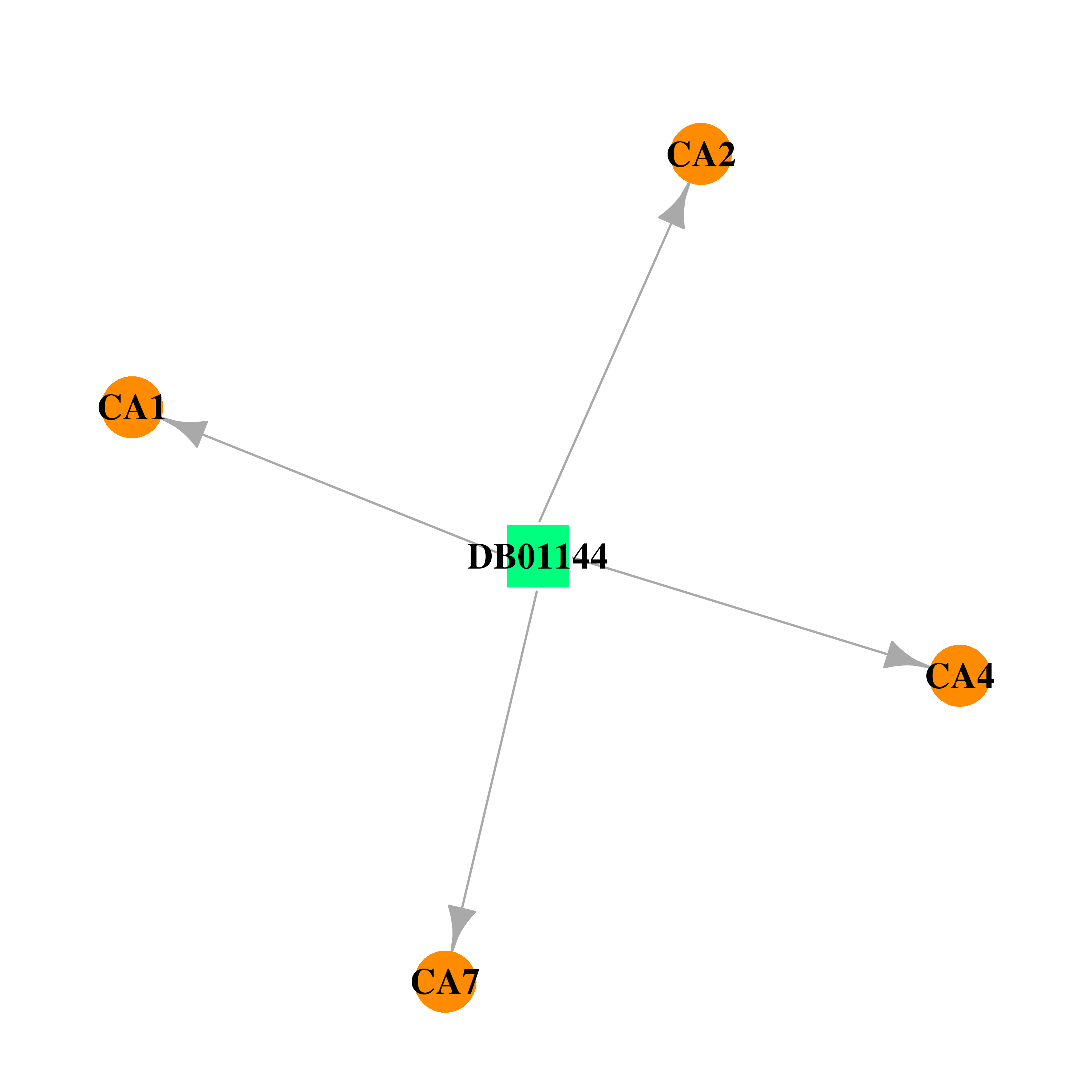 |  |
| DB01194 | carbonic anhydrase II | approved | Brinzolamide |  |  |
| DB01325 | carbonic anhydrase II | approved | Quinethazone |  |  |
| DB01671 | carbonic anhydrase II | experimental | 4-(Hydroxymercury)Benzoic Acid |  |  |
| DB01748 | carbonic anhydrase II | experimental | N-Benzyl-4-Sulfamoyl-Benzamide |  |  |
| DB01784 | carbonic anhydrase II | experimental | 4-Flourobenzenesulfonamide |  |  |
| DB01942 | carbonic anhydrase II | experimental | Formic Acid |  | 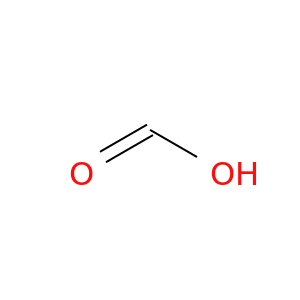 |
| DB01964 | carbonic anhydrase II | experimental | AL5424 |  | 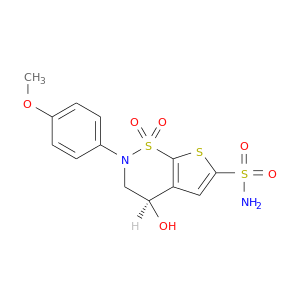 |
| DB02069 | carbonic anhydrase II | experimental | N-(2-Flouro-Benzyl)-4-Sulfamoyl-Benzamide |  |  |
| DB02087 | carbonic anhydrase II | experimental | 3,5-Difluorobenzenesulfonamide |  |  |
| DB02220 | carbonic anhydrase II | experimental | Al7089a |  |  |
| DB02221 | carbonic anhydrase II | experimental | 4-(Aminosulfonyl)-N-[(2,4,6-Trifluorophenyl)Methyl]-Benzamide |  |  |
| DB02292 | carbonic anhydrase II | experimental | 6-Oxo-8,9,10,11-Tetrahydro-7h-Cyclohepta[C][1]Benzopyran-3-O-Sulfamate |  |  |
| DB02429 | carbonic anhydrase II | experimental | 4-(Aminosulfonyl)-N-[(4-Fluorophenyl)Methyl]-Benzamide |  |  |
| DB02479 | carbonic anhydrase II | experimental | (R)-N-(3-Indol-1-Yl-2-Methyl-Propyl)-4-Sulfamoyl-Benzamide |  |  |
| DB02535 | carbonic anhydrase II | experimental | Aminodi(Ethyloxy)Ethylaminocarbonylbenzenesulfonamide |  |  |
| DB02602 | carbonic anhydrase II | experimental | AL7182 |  |  |
| DB02610 | carbonic anhydrase II | experimental | N-(2,3,4,5,6-Pentaflouro-Benzyl)-4-Sulfamoyl-Benzamide |  |  |
| DB02679 | carbonic anhydrase II | experimental | Cyanamide |  |  |
| DB02861 | carbonic anhydrase II | experimental | 4-(Aminosulfonyl)-N-[(3,4,5-Trifluorophenyl)Methyl]-Benzamide |  | 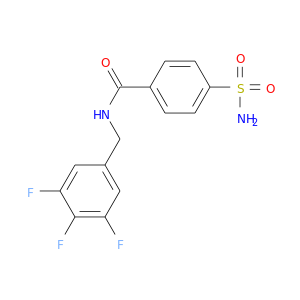 |
| DB02866 | carbonic anhydrase II | experimental | Dansylamide |  | 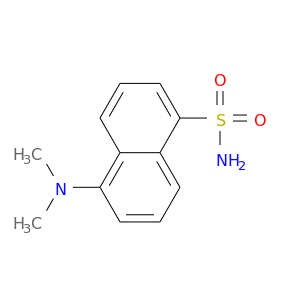 |
| DB02894 | carbonic anhydrase II | experimental | Sulfamic Acid 2,3-O-(1-Methylethylidene)-4,5-O-Sulfonyl-Beta-Fructopyranose Ester |  | 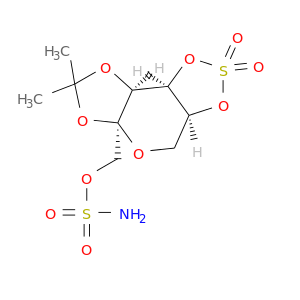 |
| DB02986 | carbonic anhydrase II | experimental | N-(2-Thienylmethyl)-2,5-Thiophenedisulfonamide |  | 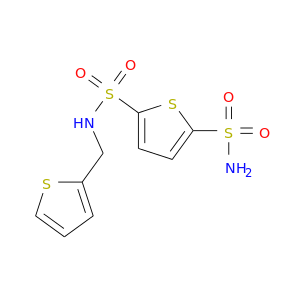 |
| DB03039 | carbonic anhydrase II | experimental | 4-(Aminosulfonyl)-N-[(2,5-Difluorophenyl)Methyl]-Benzamide |  |  |
| DB03221 | carbonic anhydrase II | experimental | AL7099A |  |  |
| DB03262 | carbonic anhydrase II | experimental | Al-6619, [2h-Thieno[3,2-E]-1,2-Thiazine-6-Sulfonamide,2-(3-Hydroxyphenyl)-3-(4-Morpholinyl)-, 1,1-Dioxide] |  |  |
| DB03270 | carbonic anhydrase II | experimental | 2,6-Difluorobenzenesulfonamide |  |  |
| DB03294 | carbonic anhydrase II | experimental | 1-Methyl-3-Oxo-1,3-Dihydro-Benzo[C]Isothiazole-5-Sulfonic Acid Amide | 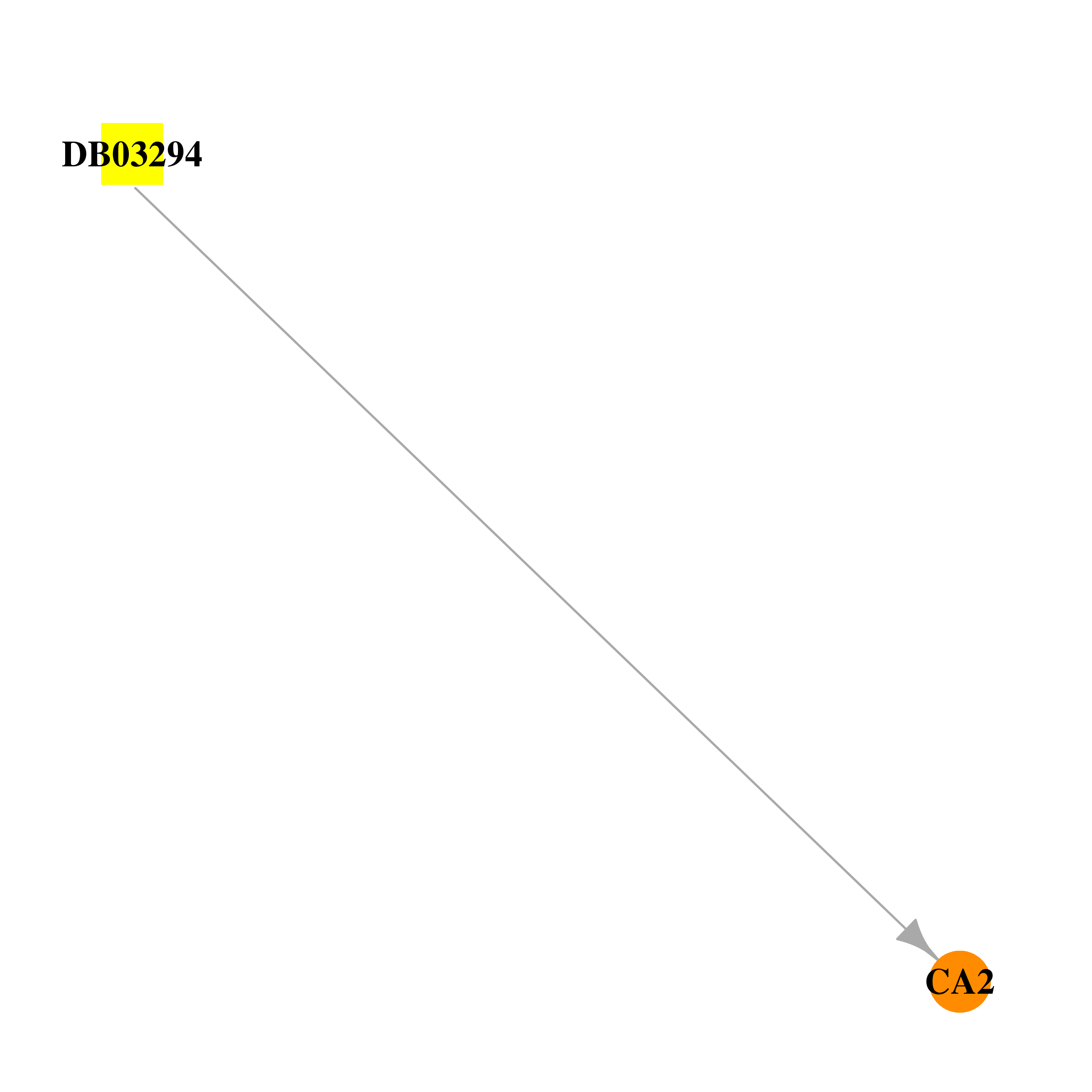 |  |
| DB03333 | carbonic anhydrase II | experimental | (4-sulfamoyl-phenyl)-thiocarbamic acid O-(2-thiophen-3-yl-ethyl) ester | 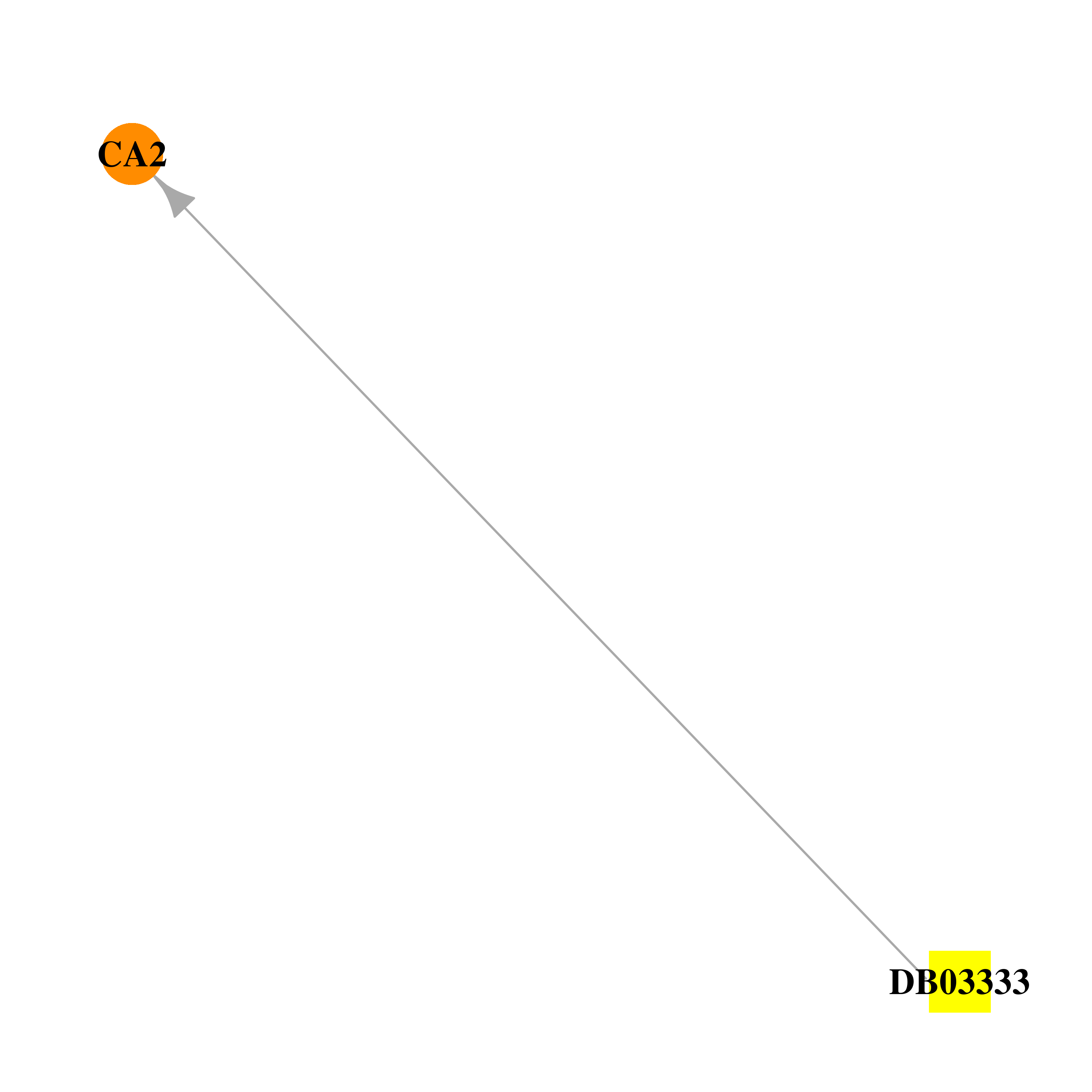 |  |
| DB03385 | carbonic anhydrase II | experimental | 4-Methylimidazole |  |  |
| DB03526 | carbonic anhydrase II | experimental | AL5927 |  |  |
| DB03594 | carbonic anhydrase II | experimental | 1,2,4-Triazole |  |  |
| DB03596 | carbonic anhydrase II | experimental | N-[2-(1h-Indol-5-Yl)-Butyl]-4-Sulfamoyl-Benzamide |  |  |
| DB03598 | carbonic anhydrase II | experimental | Al-6629, [2h-Thieno[3,2-E]-1,2-Thiazine-6-Sulfonamide,2-(3-Methoxyphenyl)-3-(4-Morpholinyl)-, 1,1-Dioxide] |  | 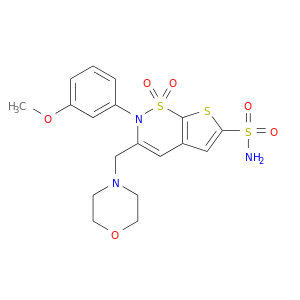 |
| DB03674 | carbonic anhydrase II | experimental | Methyl Mercury Ion |  | 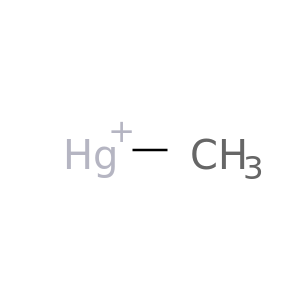 |
| DB03697 | carbonic anhydrase II | experimental | 4-Sulfonamide-[1-(4-Aminobutane)]Benzamide |  |  |
| DB03844 | carbonic anhydrase II | experimental | N-(2,6-Diflouro-Benzyl)-4-Sulfamoyl-Benzamide |  |  |
| DB03877 | carbonic anhydrase II | experimental | AL4623 |  |  |
| DB03904 | carbonic anhydrase II | experimental | Urea |  | 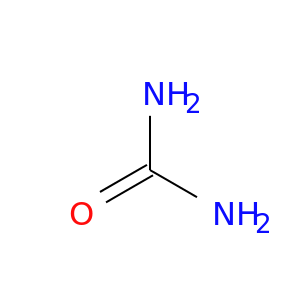 |
| DB03950 | carbonic anhydrase II | experimental | (S)-N-(3-Indol-1-Yl-2-Methyl-Propyl)-4-Sulfamoyl-Benzamide |  | 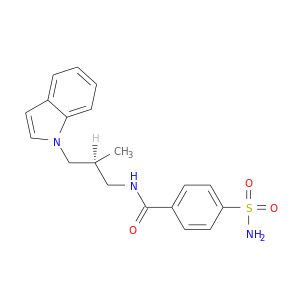 |
| DB03975 | carbonic anhydrase II | experimental | Mercuribenzoic Acid |  |  |
| DB04002 | carbonic anhydrase II | experimental | 4-Sulfonamide-[4-(Thiomethylaminobutane)]Benzamide |  |  |
| DB04081 | carbonic anhydrase II | experimental | (4s-Trans)-4-(Methylamino)-5,6-Dihydro-6-Methyl-4h-Thieno(2,3-B)Thiopyran-2-Sulfonamide-7,7-Dioxide |  |  |
| DB04089 | carbonic anhydrase II | experimental | AL5300 |  |  |
| DB04180 | carbonic anhydrase II | experimental | 4-(Aminosulfonyl)-N-[(2,4-Difluorophenyl)Methyl]-Benzamide |  |  |
| DB04203 | carbonic anhydrase II | experimental | 3-Mercuri-4-Aminobenzenesulfonamide |  |  |
| DB04371 | carbonic anhydrase II | experimental | AL6528 |  |  |
| DB04394 | carbonic anhydrase II | experimental | 3-Nitro-4-(2-Oxo-Pyrrolidin-1-Yl)-Benzenesulfonamide |  |  |
| DB04549 | carbonic anhydrase II | experimental | 4-(Aminosulfonyl)-N-[(2,3,4-Trifluorophenyl)Methyl]-Benzamide |  |  |
| DB04600 | carbonic anhydrase II | experimental | 4-[(3-BROMO-4-O-SULFAMOYLBENZYL)(4-CYANOPHENYL)AMINO]-4H-[1,2,4]-TRIAZOLE |  |  |
| DB04601 | carbonic anhydrase II | experimental | 4-[(4-O-SULFAMOYLBENZYL)(4-CYANOPHENYL)AMINO]-4H-[1,2,4]-TRIAZOLE |  |  |
| DB04763 | carbonic anhydrase II | experimental | 1-N-(4-SULFAMOYLPHENYL-ETHYL)-2,4,6-TRIMETHYLPYRIDINIUM |  |  |
| DB06891 | carbonic anhydrase II | experimental | 5-{[(4-AMINO-3-CHLORO-5-FLUOROPHENYL)SULFONYL]AMINO}-1,3,4-THIADIAZOLE-2-SULFONAMIDE |  | 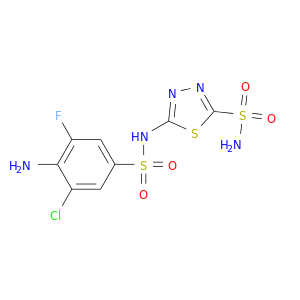 |
| DB06954 | carbonic anhydrase II | experimental | 2-(cycloheptylmethyl)-1,1-dioxido-1-benzothiophen-6-yl sulfamate |  | 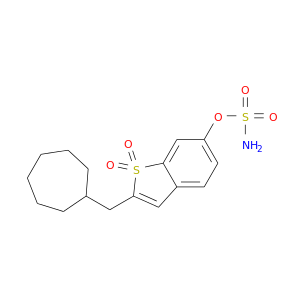 |
| DB07048 | carbonic anhydrase II | experimental | N-[(2R)-5-(aminosulfonyl)-2,3-dihydro-1H-inden-2-yl]-2-propylpentanamide | 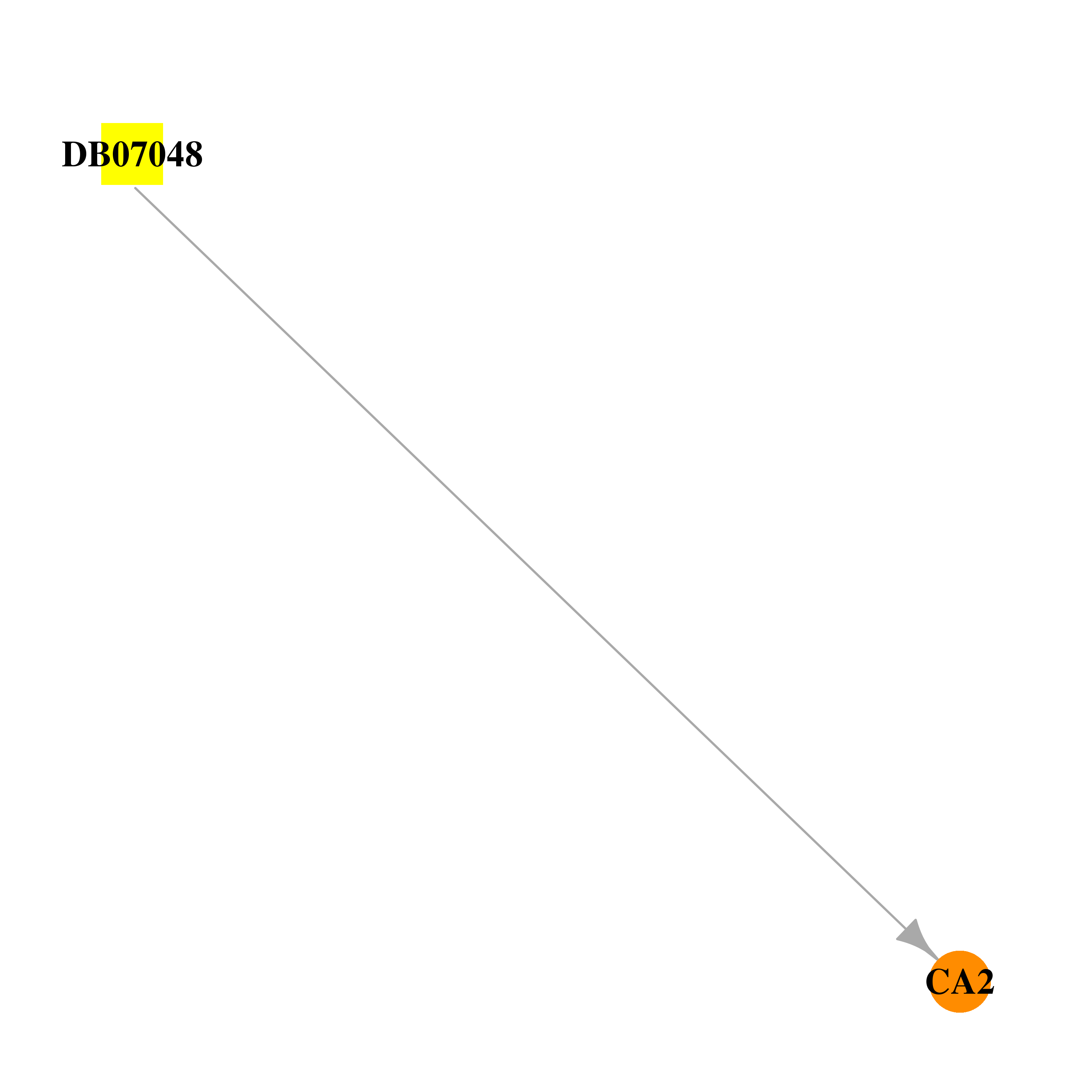 | 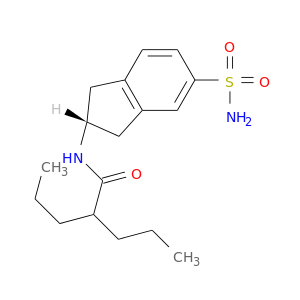 |
| DB07050 | carbonic anhydrase II | experimental | 5-[(phenylsulfonyl)amino]-1,3,4-thiadiazole-2-sulfonamide |  |  |
| DB07363 | carbonic anhydrase II | experimental | THIOPHENE-2,5-DISULFONIC ACID 2-AMIDE-5-(4-METHYL-BENZYLAMIDE) |  |  |
| DB07467 | carbonic anhydrase II | experimental | 4-chloro-N-[(2S)-2-methyl-2,3-dihydro-1H-indol-1-yl]-3-sulfamoylbenzamide |  |  |
| DB07476 | carbonic anhydrase II | experimental | N-[4-(AMINOSULFONYL)PHENYL]-2-MERCAPTOBENZAMIDE |  |  |
| DB07596 | carbonic anhydrase II | experimental | (17beta)-17-(cyanomethyl)-2-methoxyestra-1(10),2,4-trien-3-yl sulfamate |  |  |
| DB07632 | carbonic anhydrase II | experimental | 5-(2-chlorophenyl)-1,3,4-thiadiazole-2-sulfonamide |  |  |
| DB07710 | carbonic anhydrase II | experimental | PHENYLALANYLAMINODI(ETHYLOXY)ETHYL BENZENESULFONAMIDEAMINOCARBONYLBENZENESULFONAMIDE |  |  |
| DB07742 | carbonic anhydrase II | experimental | N-(2,3-DIFLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE |  |  |
| DB08046 | carbonic anhydrase II | experimental | 2-chloro-5-[(1S)-1-hydroxy-3-oxo-2H-isoindol-1-yl]benzenesulfonamide |  |  |
| DB08083 | carbonic anhydrase II | experimental | 2-(1,3-thiazol-4-yl)-1H-benzimidazole-5-sulfonamide |  |  |
| DB08155 | carbonic anhydrase II | experimental | N-{2-[4-(AMINOSULFONYL)PHENYL]ETHYL}ACETAMIDE |  |  |
| DB08156 | carbonic anhydrase II | experimental | 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID |  |  |
| DB08157 | carbonic anhydrase II | experimental | ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE |  |  |
| DB08165 | carbonic anhydrase II | experimental | indane-5-sulfonamide |  |  |
| DB08202 | carbonic anhydrase II | experimental | 4-({[(4-METHYLPIPERAZIN-1-YL)AMINO]CARBONOTHIOYL}AMINO)BENZENESULFONAMIDE |  |  |
| DB08301 | carbonic anhydrase II | experimental | N-({[4-(AMINOSULFONYL)PHENYL]AMINO}CARBONYL)-4-METHYLBENZENESULFONAMIDE |  |  |
| DB08329 | carbonic anhydrase II | experimental | SULTHIAME |  |  |
| DB08416 | carbonic anhydrase II | experimental | (9BETA,13ALPHA,14BETA,17ALPHA)-2-METHOXYESTRA-1,3,5(10)-TRIENE-3,17-DIYL DISULFAMATE |  |  |
| DB08418 | carbonic anhydrase II | experimental | (4aS,4bR,10bS,12aS)-12a-methyl-1,3-dioxo-2-(pyridin-3-ylmethyl)-1,2,3,4,4a,4b,5,6,10b,11,12,12a-dodecahydronaphtho[2,1-f]isoquinolin-8-yl sulfamate |  |  |
| DB08645 | carbonic anhydrase II | experimental | 6-CHLORO-3-(DICHLOROMETHYL)-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE-7-SULFONAMIDE 1,1-DIOXIDE |  |  |
| DB08659 | carbonic anhydrase II | experimental | 2-(hydrazinocarbonyl)-3-phenyl-1H-indole-5-sulfonamide |  |  |
| DB08765 | carbonic anhydrase II | experimental | 6-HYDROXY-1,3-BENZOTHIAZOLE-2-SULFONAMIDE | 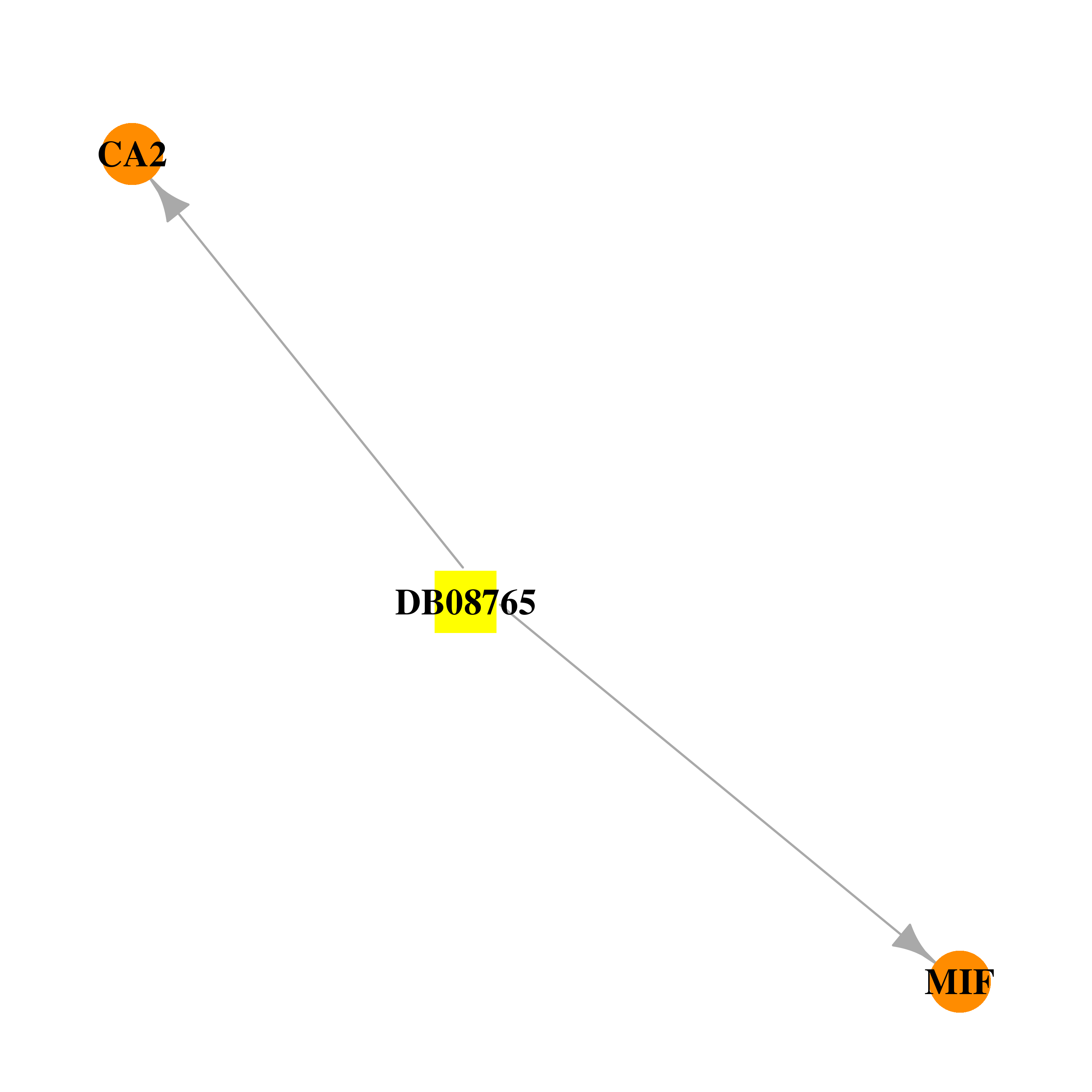 |  |
| DB08782 | carbonic anhydrase II | experimental | 4-(2-AMINOETHYL)BENZENESULFONAMIDE |  |  |
| DB00259 | carbonic anhydrase II | approved | Sulfanilamide | 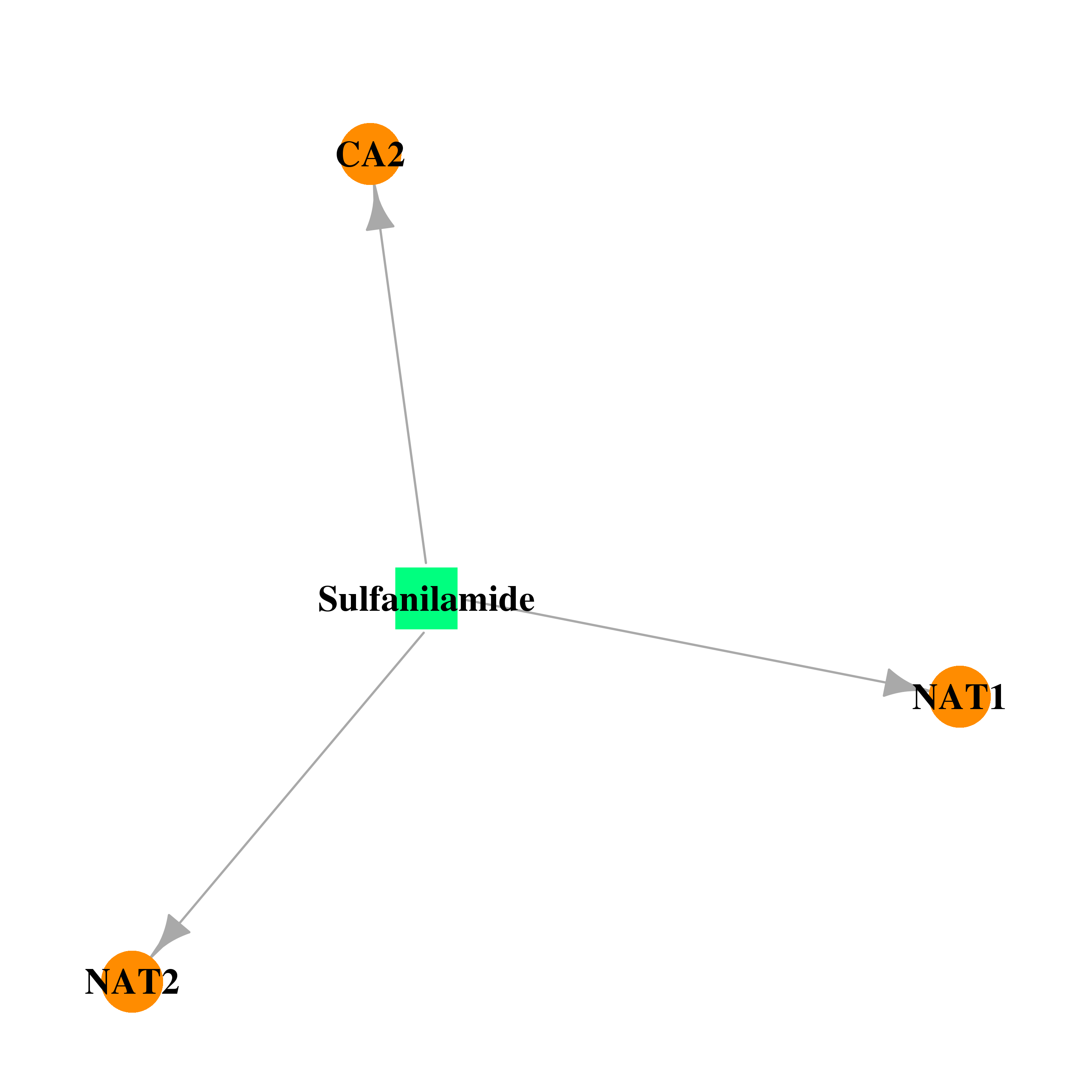 |  |
| Top |
| Cross referenced IDs for CA2 |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |