|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for CALM1 |
| Top |
| Phenotypic Information for CALM1(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: CALM1 |
| Familial Cancer Database: CALM1 | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
| . | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Oncogene 1 | Significant driver gene in | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| REACTOME_METABOLISM_OF_CARBOHYDRATES REACTOME_GLUCOSE_METABOLISM | |
| OMIM | |
| Orphanet | |
| Disease | KEGG Disease: CALM1 |
| MedGen: CALM1 (Human Medical Genetics with Condition) | |
| ClinVar: CALM1 | |
| Phenotype | MGI: CALM1 (International Mouse Phenotyping Consortium) |
| PhenomicDB: CALM1 | |
| Mutations for CALM1 |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| - Statistics for Tissue and Mutation type | Top |
 |
| - For Inter-chromosomal Variations |
| * Inter-chromosomal variantions includes 'interchromosomal amplicon to amplicon', 'interchromosomal amplicon to non-amplified dna', 'interchromosomal insertion', 'Interchromosomal unknown type'. |
 |
| - For Intra-chromosomal Variations |
| * Intra-chromosomal variantions includes 'intrachromosomal amplicon to amplicon', 'intrachromosomal amplicon to non-amplified dna', 'intrachromosomal deletion', 'intrachromosomal fold-back inversion', 'intrachromosomal inversion', 'intrachromosomal tandem duplication', 'Intrachromosomal unknown type', 'intrachromosomal with inverted orientation', 'intrachromosomal with non-inverted orientation'. |
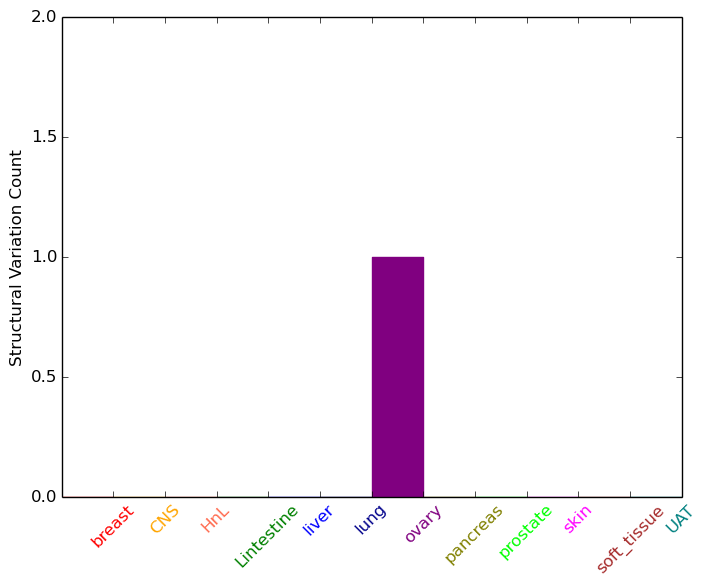 |
| Sample | Symbol_a | Chr_a | Start_a | End_a | Symbol_b | Chr_b | Start_b | End_b |
| ovary | CALM1 | chr14 | 90867961 | 90867981 | CALM1 | chr14 | 90868518 | 90868538 |
| cf) Tissue number; Tissue name (1;Breast, 2;Central_nervous_system, 3;Haematopoietic_and_lymphoid_tissue, 4;Large_intestine, 5;Liver, 6;Lung, 7;Ovary, 8;Pancreas, 9;Prostate, 10;Skin, 11;Soft_tissue, 12;Upper_aerodigestive_tract) |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows CALM1 related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| BF898266 | CALM1 | 11 | 69 | 14 | 90863474 | 90863532 | MINK1 | 66 | 270 | 17 | 4791027 | 4793840 | |
| AI141454 | DCXR | 8 | 305 | 17 | 79993757 | 79994181 | CALM1 | 302 | 372 | 14 | 90873731 | 90873801 | |
| BF218294 | CALM1 | 2 | 235 | 14 | 90872253 | 90872488 | PSME3 | 226 | 318 | 17 | 40995163 | 40995255 | |
| BE765928 | IRF2BP2 | 11 | 185 | 1 | 234741708 | 234741882 | CALM1 | 176 | 266 | 14 | 90873556 | 90873646 | |
| BE813514 | CALM1 | 1 | 391 | 14 | 90873852 | 90874245 | MGP | 385 | 468 | 12 | 15034952 | 15035035 | |
| AW993959 | CALM1 | 1 | 393 | 14 | 90873852 | 90874245 | MGP | 387 | 470 | 12 | 15034952 | 15035035 | |
| AA496229 | CALM1 | 1 | 152 | 14 | 90872502 | 90872653 | UNC5CL | 152 | 517 | 6 | 41029533 | 41032107 | |
| BE813702 | CALM1 | 1 | 387 | 14 | 90873861 | 90874245 | MGP | 381 | 464 | 12 | 15034952 | 15035035 | |
| AW993976 | CALM1 | 1 | 389 | 14 | 90873852 | 90874245 | MGP | 383 | 466 | 12 | 15034952 | 15035035 | |
| CD366766 | CALM1 | 1 | 154 | 14 | 90870820 | 90871147 | RALA | 155 | 545 | 7 | 39730002 | 39745749 | |
| BE813534 | CALM1 | 1 | 392 | 14 | 90873852 | 90874245 | MGP | 386 | 469 | 12 | 15034952 | 15035035 | |
| AW993963 | CALM1 | 1 | 389 | 14 | 90873852 | 90874245 | MGP | 383 | 466 | 12 | 15034952 | 15035035 | |
| AW993962 | CALM1 | 1 | 392 | 14 | 90873852 | 90874245 | MGP | 386 | 469 | 12 | 15034952 | 15035035 | |
| AW993965 | CALM1 | 1 | 390 | 14 | 90873852 | 90874245 | MGP | 384 | 467 | 12 | 15034952 | 15035035 | |
| BE813679 | CALM1 | 1 | 392 | 14 | 90873852 | 90874245 | MGP | 386 | 469 | 12 | 15034952 | 15035035 | |
| BF949860 | CALM1 | 2 | 78 | 14 | 90872966 | 90873043 | CANX | 74 | 166 | 5 | 179146676 | 179146768 | |
| CB164141 | STAU1 | 39 | 440 | 20 | 47730060 | 47730461 | CALM1 | 429 | 546 | 14 | 90871149 | 90871266 | |
| AI918711 | CALM1 | 1 | 136 | 14 | 90870820 | 90871129 | CALM1 | 128 | 284 | 14 | 90871960 | 90872116 | |
| CB161392 | STAU1 | 39 | 440 | 20 | 47730060 | 47730461 | CALM1 | 429 | 536 | 14 | 90871149 | 90871256 | |
| AA421615 | AIMP1 | 1 | 192 | 4 | 107268733 | 107268924 | CALM1 | 185 | 394 | 14 | 90871714 | 90871923 | |
| AA652945 | CALM1 | 8 | 465 | 14 | 90871437 | 90871892 | CALM1 | 461 | 553 | 14 | 90871685 | 90871777 | |
| AW502357 | CERS2 | 20 | 259 | 1 | 150937735 | 150937974 | CALM1 | 250 | 518 | 14 | 90872218 | 90872487 | |
| AA319858 | CACNA1B | 1 | 91 | 9 | 140952227 | 140952317 | CALM1 | 89 | 373 | 14 | 90866408 | 90870313 | |
| M78058 | FSCN1 | 1 | 58 | 7 | 5646011 | 5646068 | CALM1 | 59 | 438 | 14 | 90871328 | 90871706 | |
| CB250526 | CALM1 | 18 | 89 | 14 | 90871093 | 90871164 | ABI2 | 82 | 635 | 2 | 204234195 | 204234749 | |
| W26114 | CALM1 | 26 | 236 | 14 | 90873821 | 90874031 | CALM1 | 235 | 510 | 14 | 90874240 | 90874517 | |
| CF891084 | CALM1 | 18 | 89 | 14 | 90871093 | 90871164 | ABI2 | 82 | 761 | 2 | 204234068 | 204234749 | |
| Top |
| Mutation type/ Tissue ID | brca | cns | cerv | endome | haematopo | kidn | Lintest | liver | lung | ns | ovary | pancre | prost | skin | stoma | thyro | urina | |||
| Total # sample | 2 | |||||||||||||||||||
| GAIN (# sample) | 1 | |||||||||||||||||||
| LOSS (# sample) | 1 |
| cf) Tissue ID; Tissue type (1; Breast, 2; Central_nervous_system, 3; Cervix, 4; Endometrium, 5; Haematopoietic_and_lymphoid_tissue, 6; Kidney, 7; Large_intestine, 8; Liver, 9; Lung, 10; NS, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary_tract) |
| Top |
|
 |
| Top |
| Stat. for Non-Synonymous SNVs (# total SNVs=11) | (# total SNVs=4) |
 |  |
(# total SNVs=0) | (# total SNVs=0) |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr14:90870756-90870756 | p.R107C | 3 |
| chr14:90870757-90870757 | p.R107H | 2 |
| chr14:90870237-90870237 | p.L70F | 2 |
| chr14:90870246-90870246 | p.M73I | 1 |
| chr14:90867643-90867643 | p.D25D | 1 |
| chr14:90870251-90870251 | p.R75I | 1 |
| chr14:90867659-90867659 | p.K31E | 1 |
| chr14:90870298-90870298 | p.R91* | 1 |
| chr14:90867678-90867678 | p.M37T | 1 |
| chr14:90870303-90870303 | p.V92V | 1 |
| Top |
|
 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 1 | 1 | 2 | |||||||||
| # mutation | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 1 | 1 | 1 | |||||||||
| nonsynonymous SNV | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | |||||||||||
| synonymous SNV | 1 | 1 | 1 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr14:90870756 | p.R107C | 2 |
| chr14:90870246 | p.L70F | 1 |
| chr14:90870251 | p.M73I | 1 |
| chr14:90870274 | p.R75I | 1 |
| chr14:90870303 | p.E83K | 1 |
| chr14:90866423 | p.V92V | 1 |
| chr14:90870757 | p.R107H | 1 |
| chr14:90867659 | p.D134N | 1 |
| chr14:90870837 | p.N138S | 1 |
| chr14:90867746 | p.T6T | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
| Gene Expression for CALM1 |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
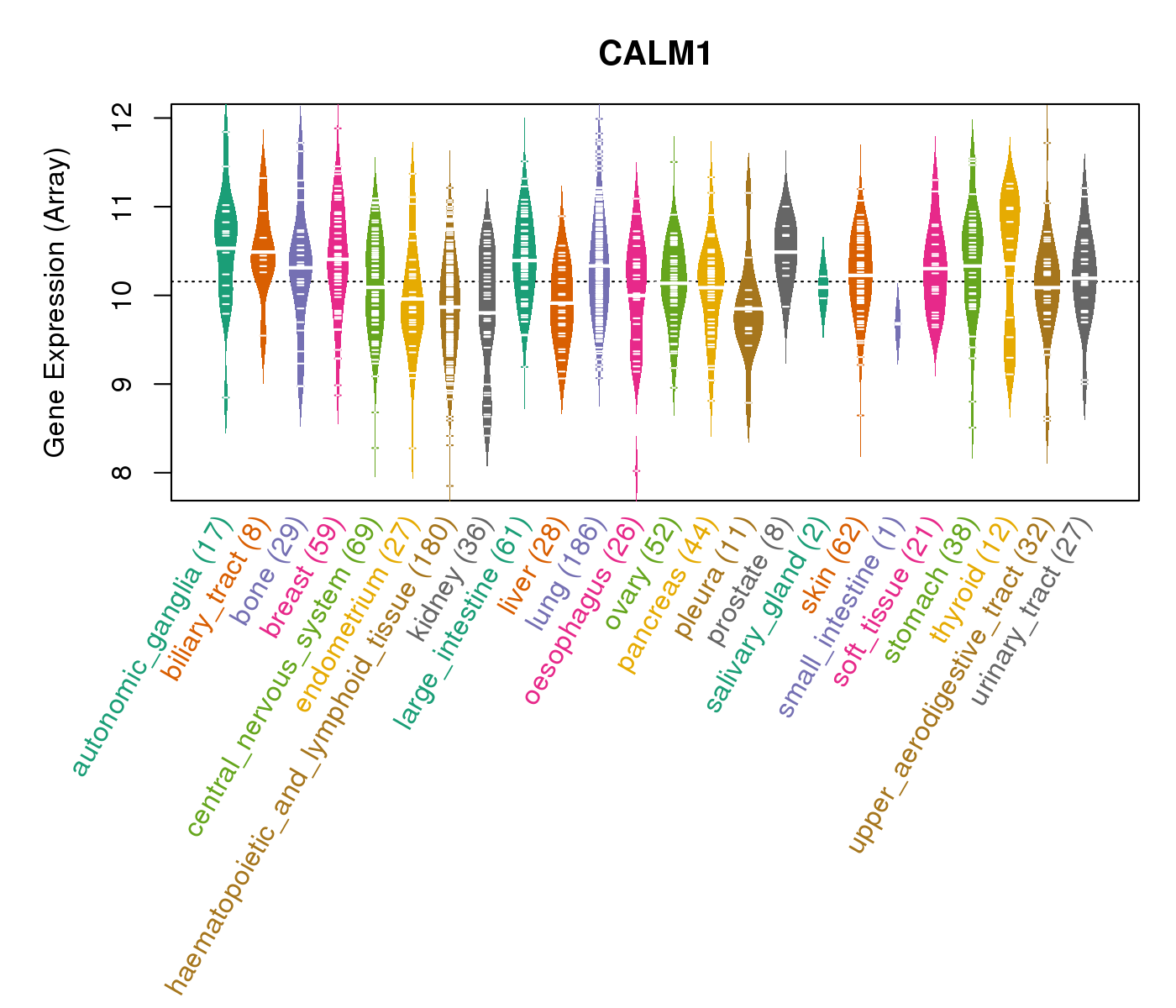 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
 |
| Top |
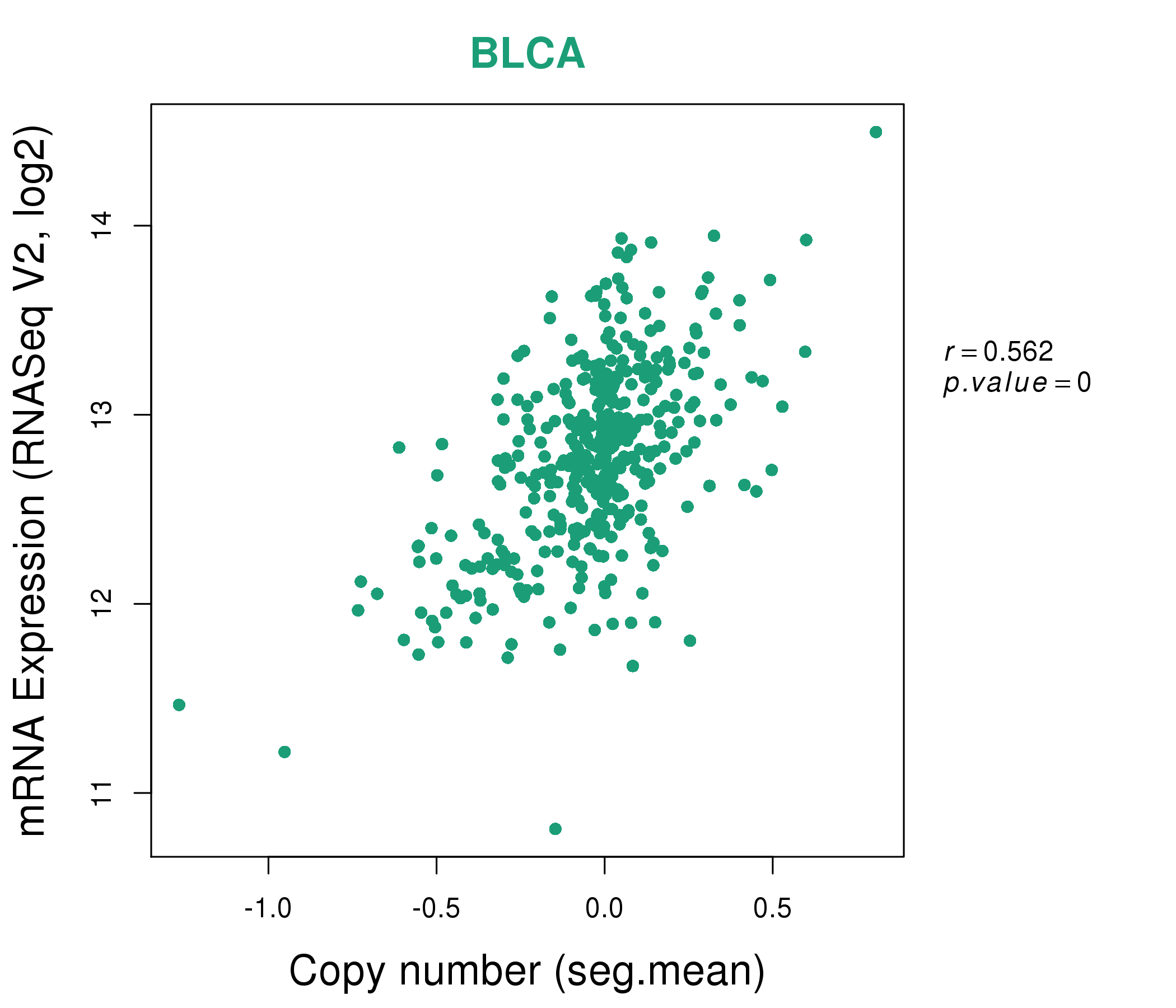
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
 |
|
 |
|
| Top |
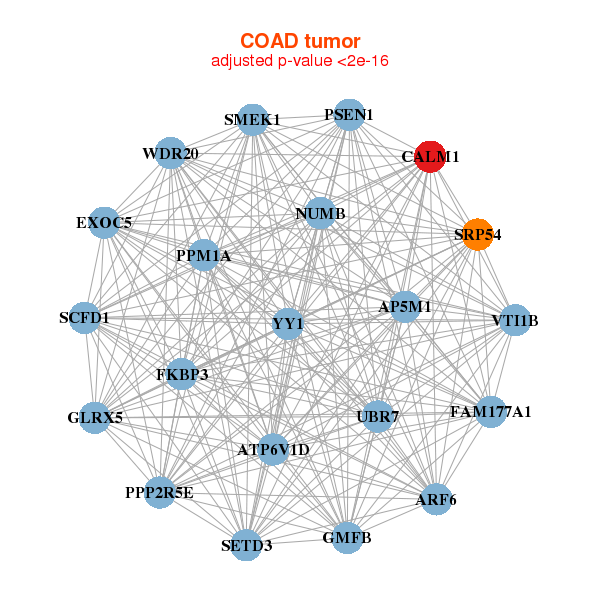
| Gene-Gene Network Information |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
 |
| ||||
| AHSA1,AKT1,ATP6V1D,C14orf1,GSKIP,CALM1,CCNK, CPSF2,DLST,EIF2B2,ERH,GOLGA5,MARK3,MED6, PPP2R5C,PSMA3,PSMC1,RDH11,SETD3,SMEK1,TTC9 | ANGPT1,ANKRD40,BCAP29,BHMT2,CALM1,CAV1,EPDR1, FAM69A,GNG2,ITGB1BP1,MAP2K1,PALMD,RHOQ,RTN4, SAR1A,SEC23A,SH3GLB1,SORBS1,TTC7B,UGP2,YWHAG | ||||
 |
| ||||
| ARF6,ATP6V1D,CALM1,EXOC5,FAM177A1,FKBP3,GLRX5, GMFB,AP5M1,NUMB,PPM1A,PPP2R5E,PSEN1,SCFD1, SETD3,SMEK1,SRP54,UBR7,VTI1B,WDR20,YY1 | ACTB,ACTR1A,CA14,CALM1,CASQ1,CDC42EP4,CKMT2, DGKG,FREM1,HRASLS,INPP5A,KCNK3,KCTD8,KLHL1, MYOM2,NTM,PCP4L1,ARHGEF26,SLC8A1,SMYD1,TUBB2A |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for CALM1 |
| DB Category | DB Name | DB's ID and Url link |
| * Gene Centered Interaction Network. |
 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB00527 | calmodulin 1 (phosphorylase kinase, delta) | approved | Dibucaine |  |  |
| DB00622 | calmodulin 1 (phosphorylase kinase, delta) | approved | Nicardipine |  |  |
| DB00623 | calmodulin 1 (phosphorylase kinase, delta) | approved | Fluphenazine |  |  |
| DB00753 | calmodulin 1 (phosphorylase kinase, delta) | approved | Isoflurane |  |  |
| DB00831 | calmodulin 1 (phosphorylase kinase, delta) | approved | Trifluoperazine |  |  |
| DB00836 | calmodulin 1 (phosphorylase kinase, delta) | approved | Loperamide |  |  |
| DB00850 | calmodulin 1 (phosphorylase kinase, delta) | approved | Perphenazine |  |  |
| DB00925 | calmodulin 1 (phosphorylase kinase, delta) | approved | Phenoxybenzamine |  |  |
| DB01023 | calmodulin 1 (phosphorylase kinase, delta) | approved; investigational | Felodipine |  |  |
| DB01065 | calmodulin 1 (phosphorylase kinase, delta) | approved; nutraceutical | Melatonin |  |  |
| DB01069 | calmodulin 1 (phosphorylase kinase, delta) | approved | Promethazine | 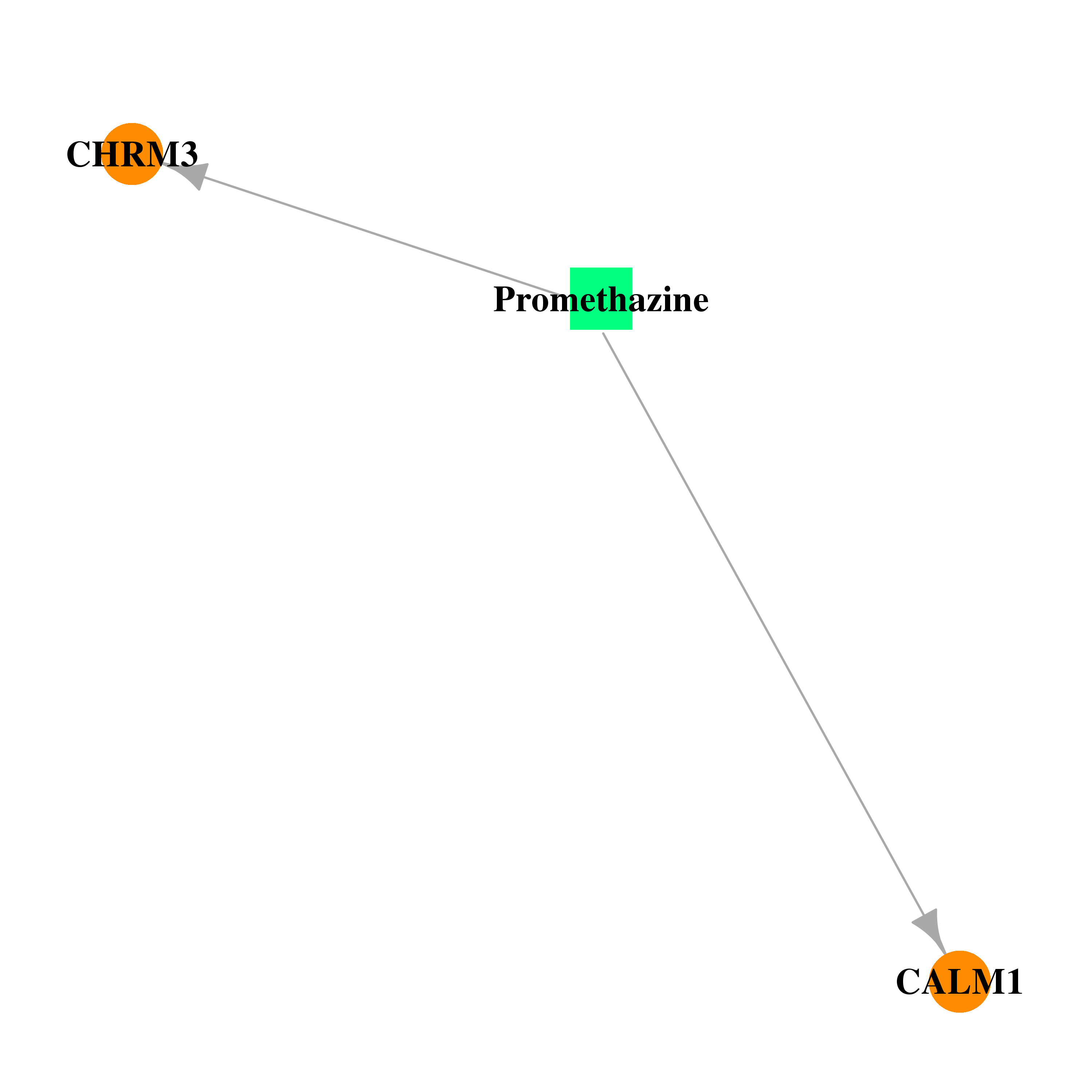 |  |
| DB01100 | calmodulin 1 (phosphorylase kinase, delta) | approved | Pimozide | 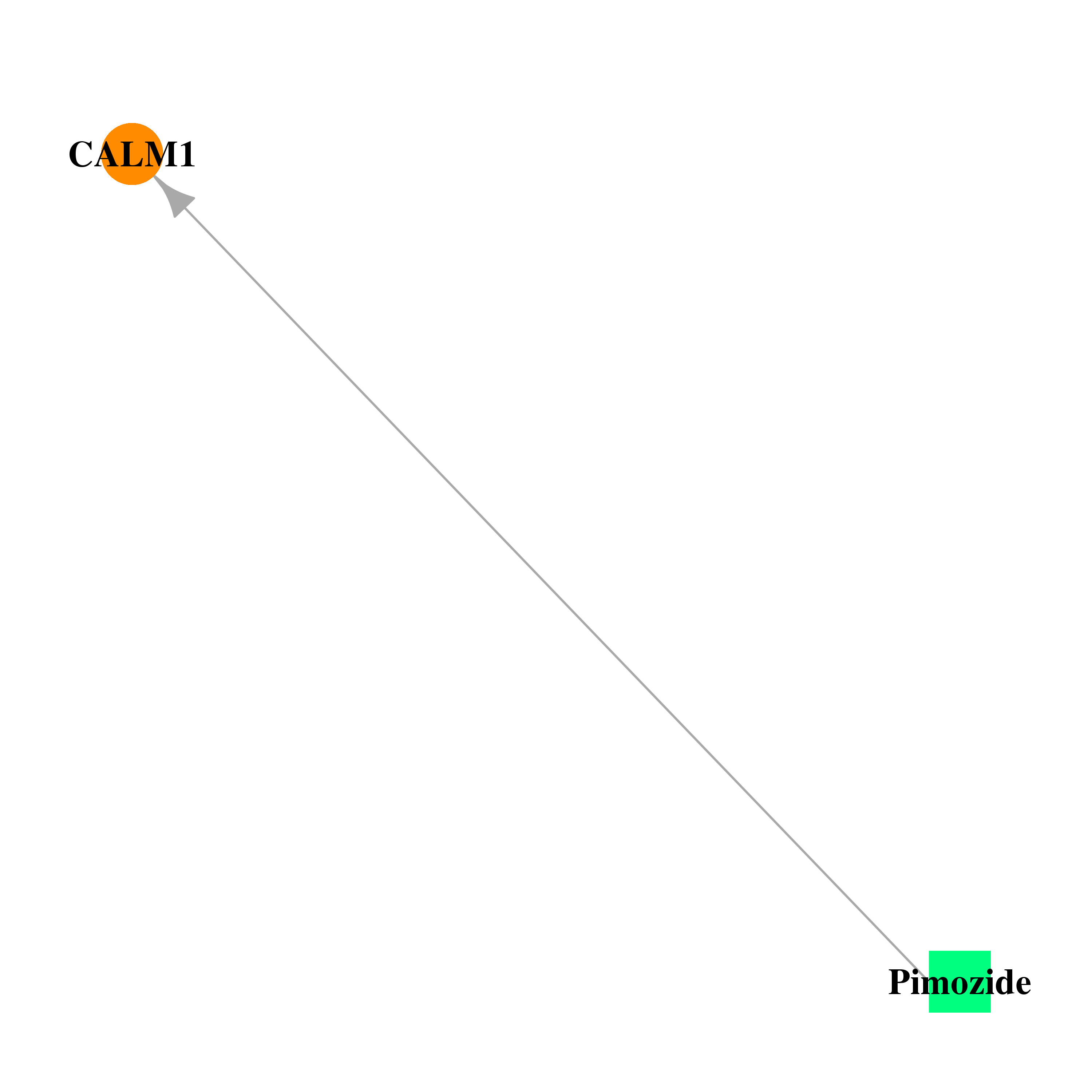 |  |
| DB01115 | calmodulin 1 (phosphorylase kinase, delta) | approved | Nifedipine | 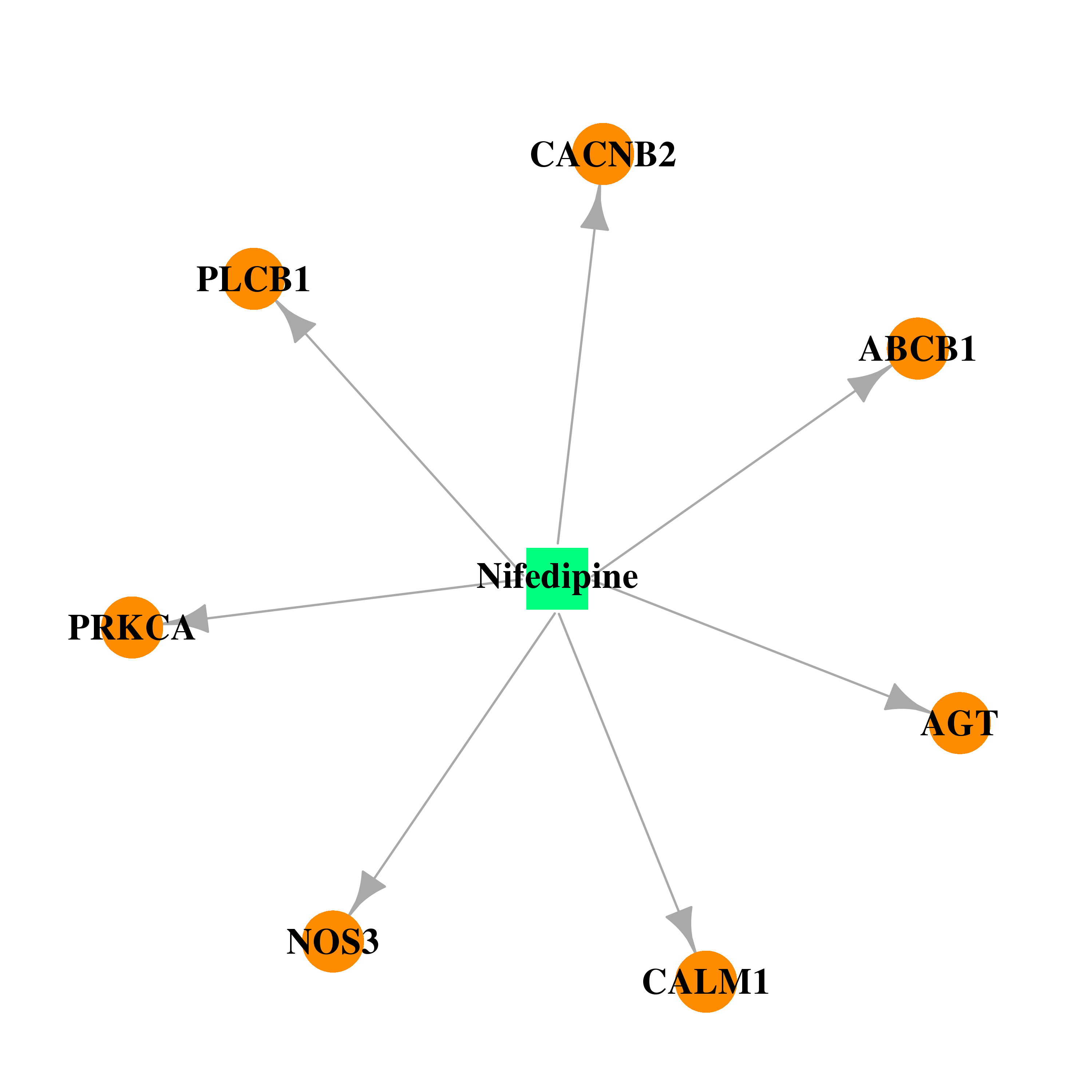 |  |
| DB01244 | calmodulin 1 (phosphorylase kinase, delta) | approved | Bepridil | 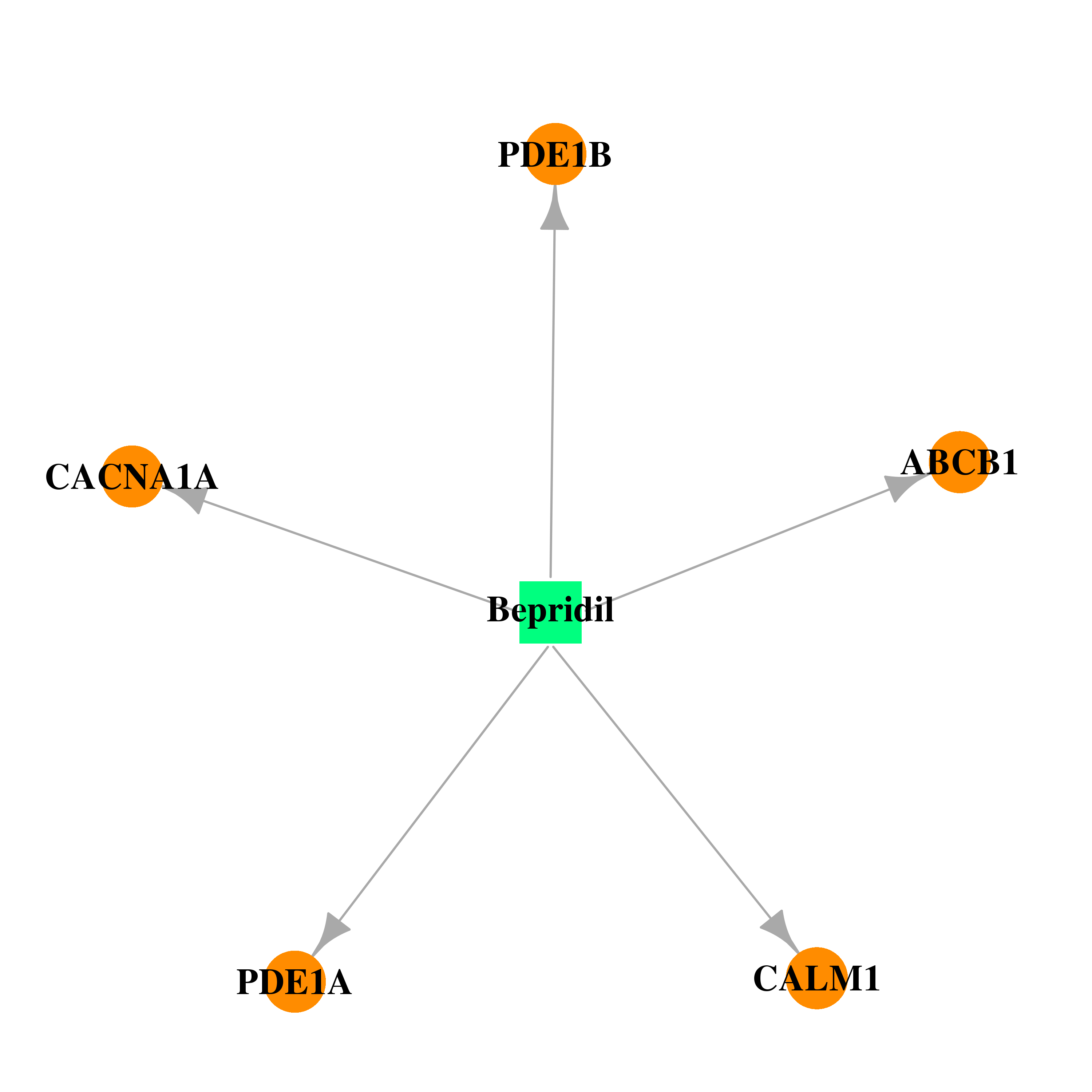 |  |
| DB01429 | calmodulin 1 (phosphorylase kinase, delta) | approved | Aprindine |  |  |
| DB02868 | calmodulin 1 (phosphorylase kinase, delta) | experimental | 3''-(Beta-Chloroethyl)-2'',4''-Dioxo-3, 5''-Spiro-Oxazolidino-4-Deacetoxy-Vinblastine | 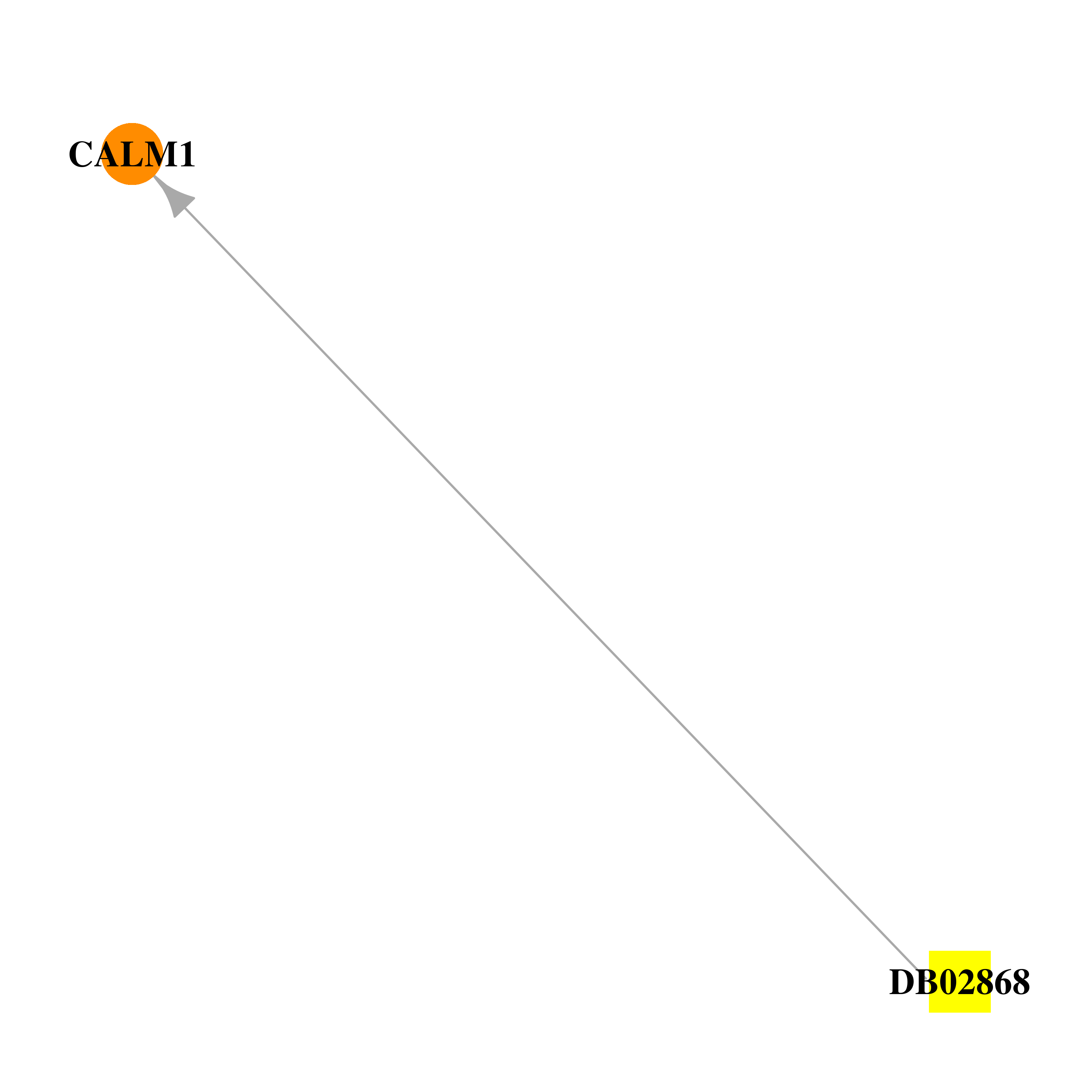 |  |
| DB03900 | calmodulin 1 (phosphorylase kinase, delta) | experimental | 2-Methyl-2-Propanol |  |  |
| DB03977 | calmodulin 1 (phosphorylase kinase, delta) | experimental | N-Trimethyllysine |  | 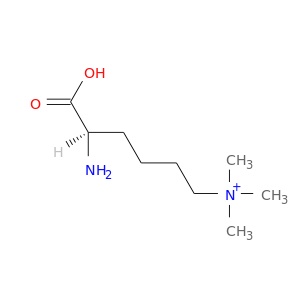 |
| DB04513 | calmodulin 1 (phosphorylase kinase, delta) | experimental | N-(6-Aminohexyl)-5-Chloro-1-Naphthalenesulfonamide |  |  |
| DB04825 | calmodulin 1 (phosphorylase kinase, delta) | withdrawn | Prenylamine |  |  |
| DB04841 | calmodulin 1 (phosphorylase kinase, delta) | approved | Flunarizine |  |  |
| DB08039 | calmodulin 1 (phosphorylase kinase, delta) | experimental | (3Z)-N,N-DIMETHYL-2-OXO-3-(4,5,6,7-TETRAHYDRO-1H-INDOL-2-YLMETHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-5-SULFONAMIDE |  |  |
| DB08231 | calmodulin 1 (phosphorylase kinase, delta) | experimental | MYRISTIC ACID |  |  |
| DB00360 | calmodulin 1 (phosphorylase kinase, delta) | approved; investigational | Tetrahydrobiopterin | 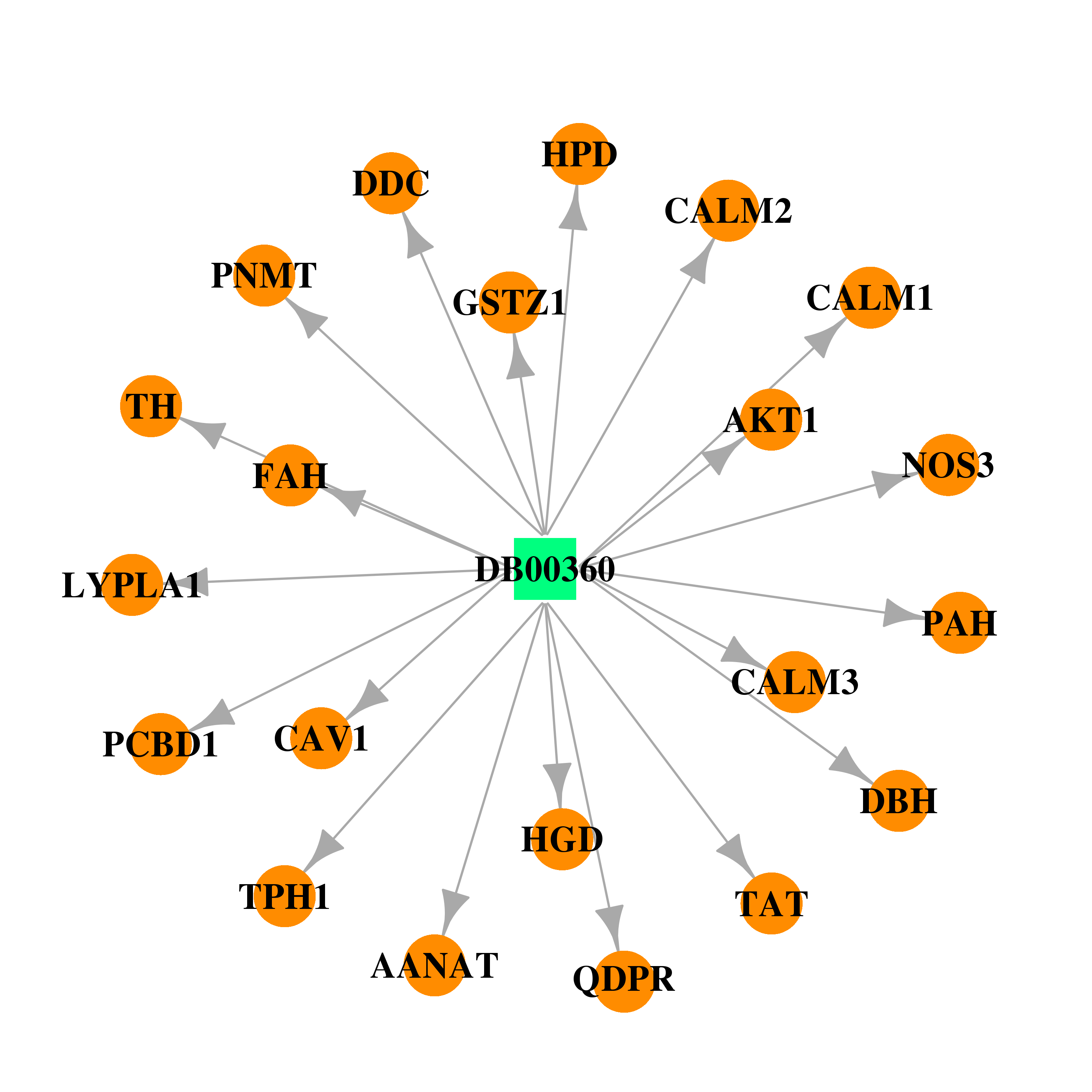 |  |
| DB00864 | calmodulin 1 (phosphorylase kinase, delta) | approved; investigational | Tacrolimus |  |  |
| DB00125 | calmodulin 1 (phosphorylase kinase, delta) | approved; nutraceutical | L-Arginine | 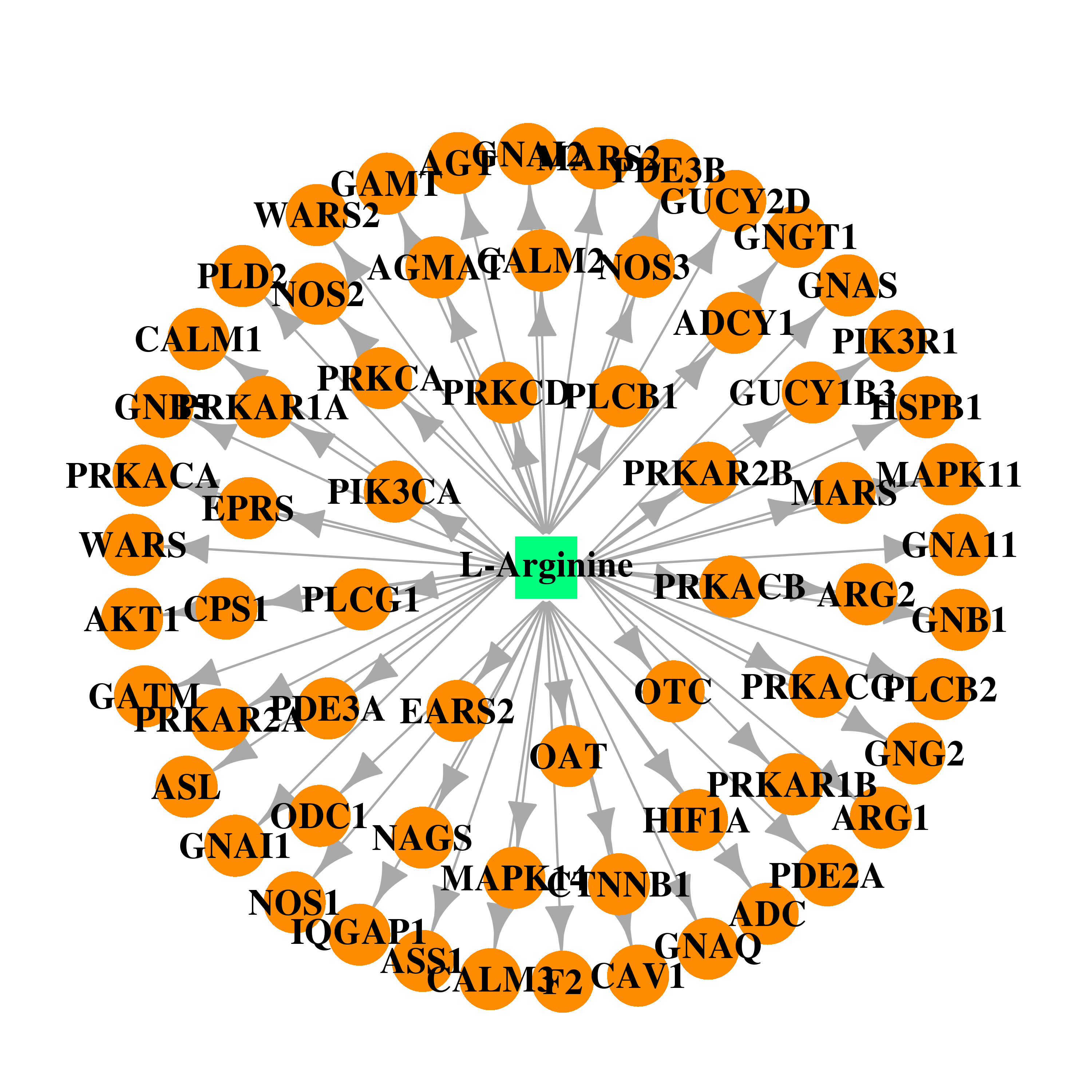 | 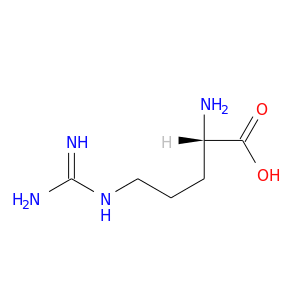 |
| DB00435 | calmodulin 1 (phosphorylase kinase, delta) | approved | Nitric Oxide | 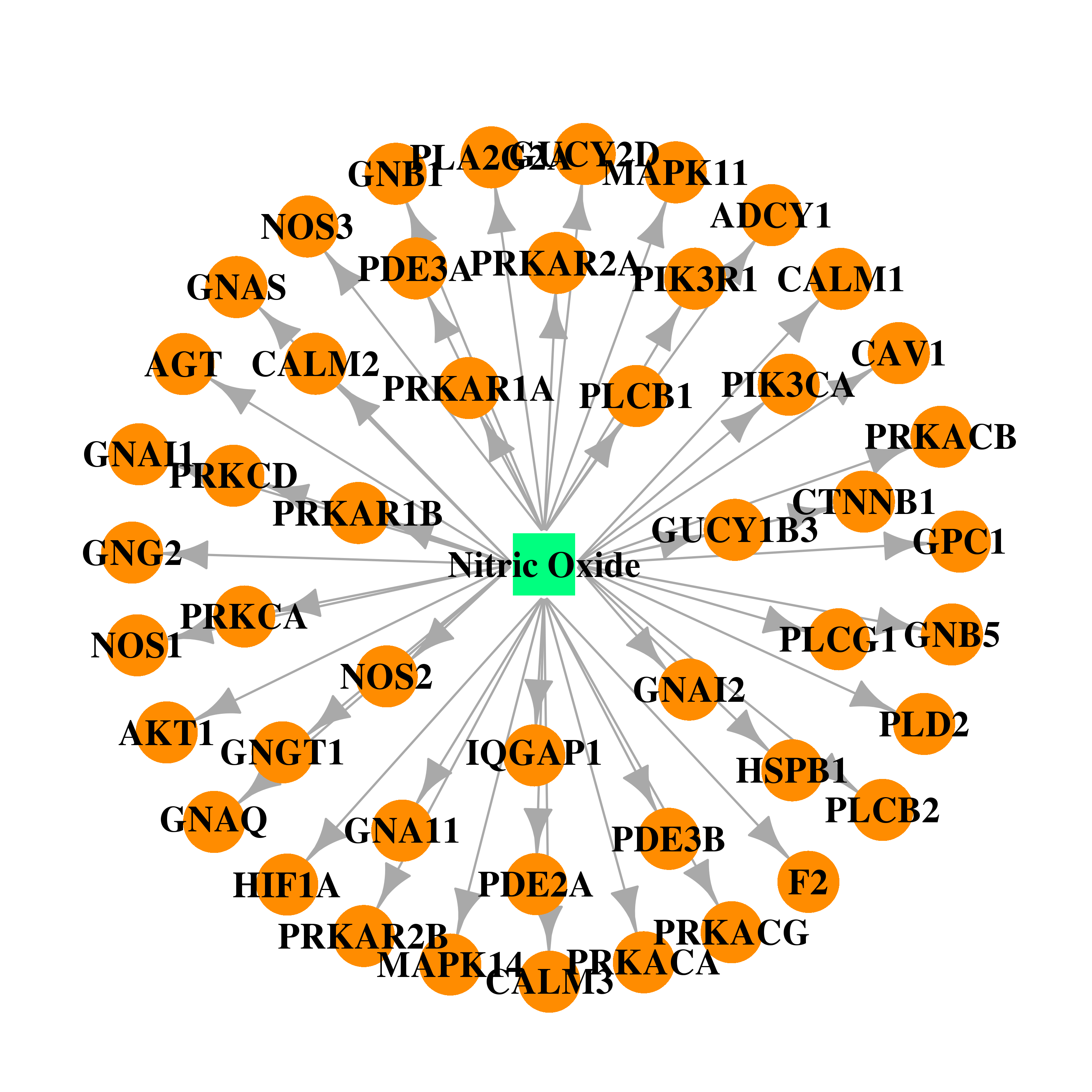 |  |
| Top |
| Cross referenced IDs for CALM1 |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |









