Network-rewiring
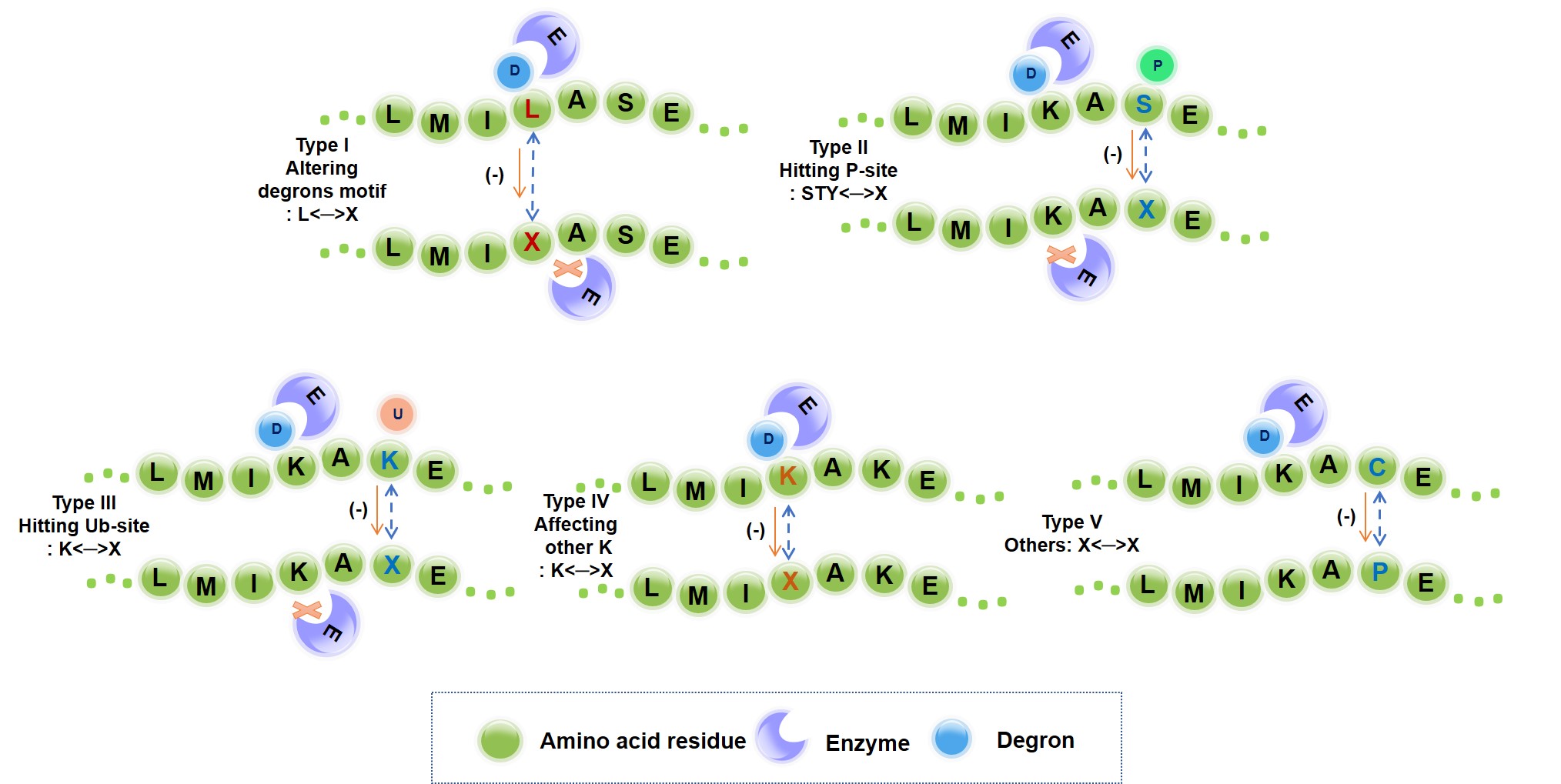
Degron muatation that destroys and perturbs UPS signals.
Network-rewiring mutationsDrug response
Pan-cancer degrome-wide drug response analysis from GDSC and CCLE database.
Browse drugsDrug resistance
Drug resistance analysis related to degrons from GDSC and CCLE database.
Browse data3D structure
Degron-1
Degron-2
Degron-3
Degron-4
Quick stats
> 18,000 new instances of degrons in the human proteome by incorporating consensus motifs with multiple degron-intrinsic structural and physicochemical features.
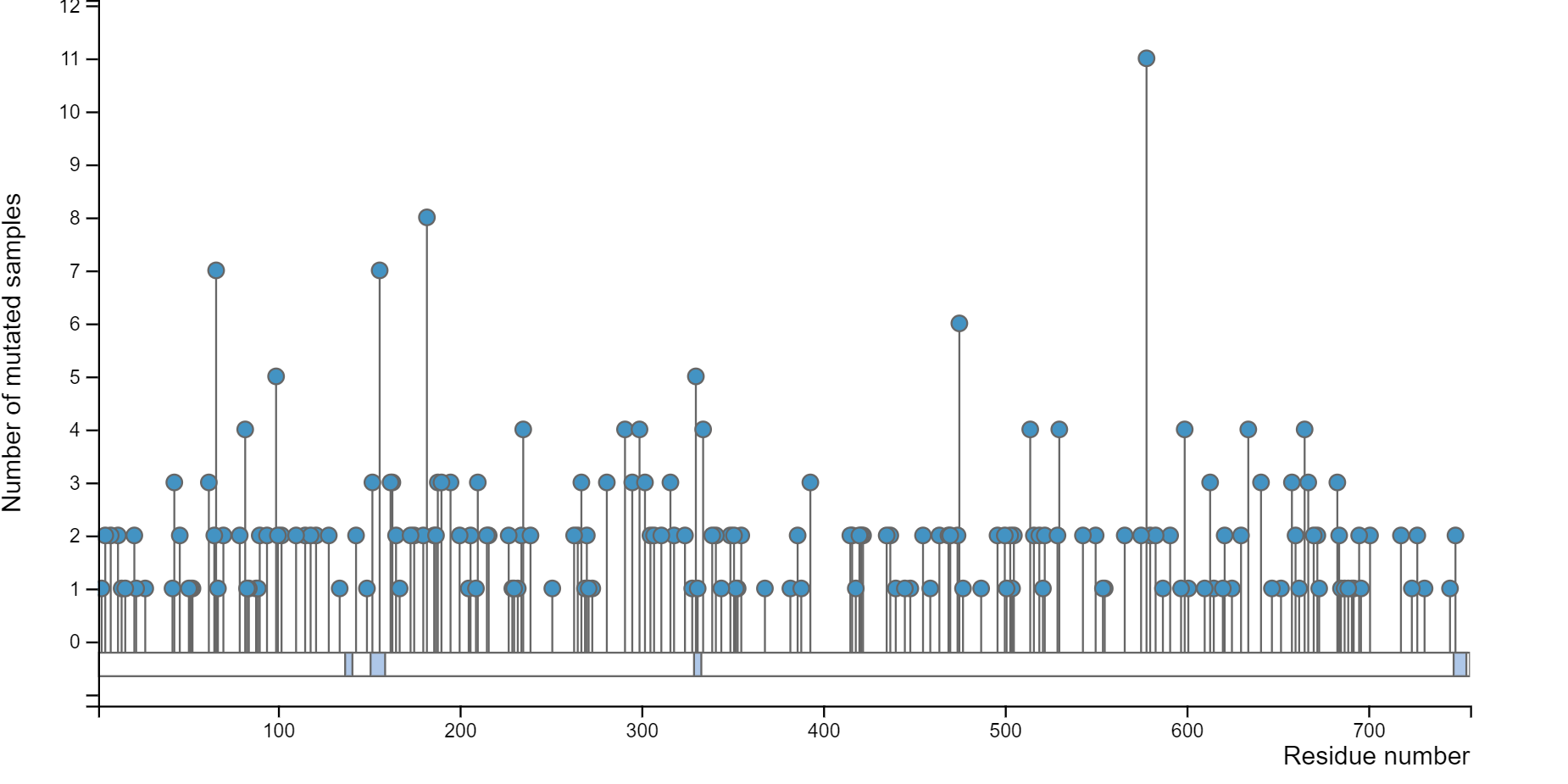
7.2 million somatic mutation records from more than 33 cancer types curated from CCLE, GDSC, TCGA, GDSC and COSMIC.
288,345 somatic mutation records on 122,503 unique amino acid positions related to degrons.
390,460 drug response records that are associated with degron proteins collected from CCLE and GDSC for pan-cancer.
378,395 and 125,994 experimentally identified protein phosphorylation and ubiquitinationsites were collected from PHOSPHOSITEPLUS.
89,318 actionable mutations were identified that directly destroys UPS signaling.
>180,000 mutations were identified that regulates UPS signaling.
7.2 million somatic mutation records from more than 33 cancer types curated from CCLE, GDSC, TCGA, GDSC and COSMIC.
288,345 somatic mutation records on 122,503 unique amino acid positions related to degrons.
390,460 drug response records that are associated with degron proteins collected from CCLE and GDSC for pan-cancer.
378,395 and 125,994 experimentally identified protein phosphorylation and ubiquitinationsites were collected from PHOSPHOSITEPLUS.
89,318 actionable mutations were identified that directly destroys UPS signaling.
>180,000 mutations were identified that regulates UPS signaling.
Updates
Mar 16, 2023: Bugs were fixed for showing the 3D structures.
Mar 12, 2023: DegronMD website and the main functions include, browsing, querying, and visualization are implemented.
Feb 10, 2023: We systematically investigated and quantified the impact of missense mutations on the functionality of a degron.
Dec 06, 2022: These mutations were then mapped to the regions of all new degron instances resulting in 288,345 somatic mutation records on 122,503 unique amino acid positions.
Nov 26, 2022: ∼7.2 million somatic mutation records were curated from five cancer-related datasets: CCLE, GDSC, TCGA, GDSC and COSMIC.
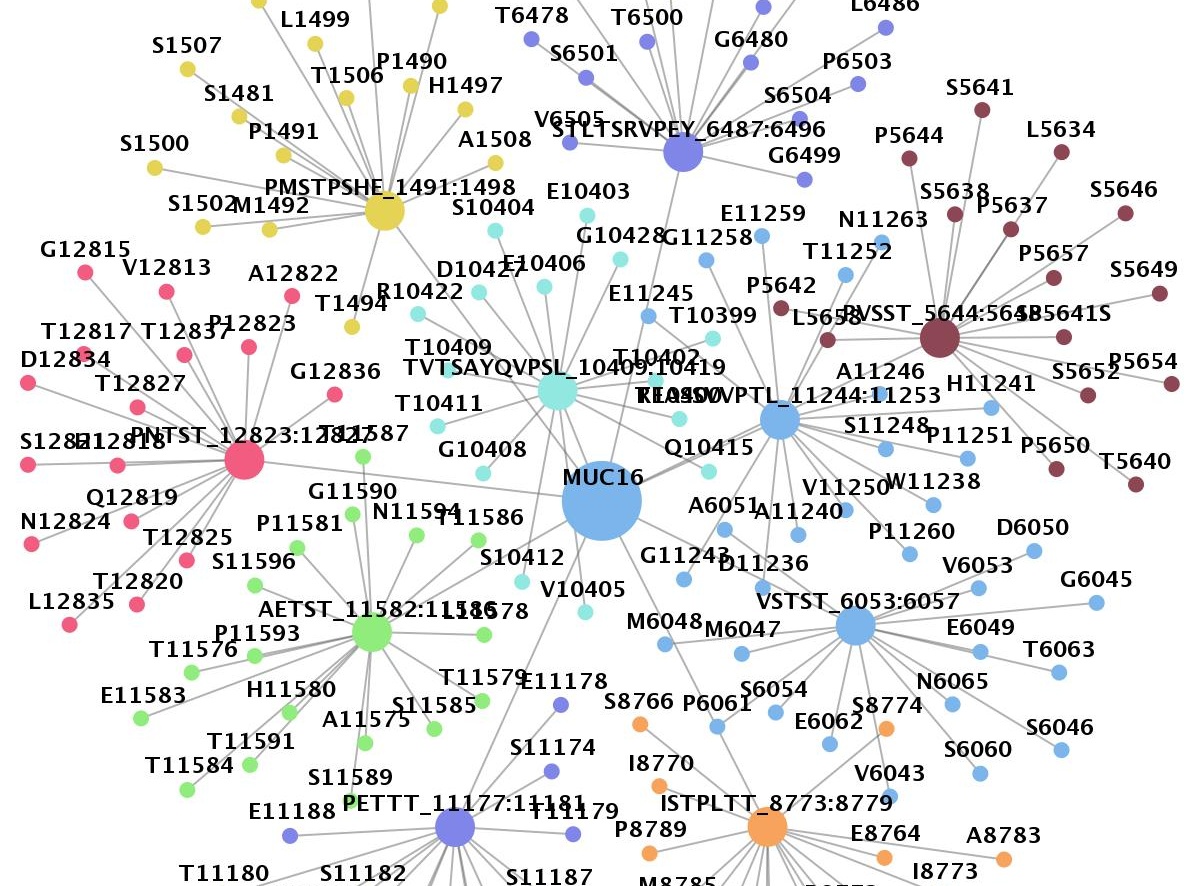
Oct 28, 2022: 18,929 new degron instances were idenfication in more than 7000 different protein.
Oct 12, 2022: A XGBoost classifier trained on known degron instances by incorporating consensus motifs with multiple degron-intrinsic structural and physicochemical features.
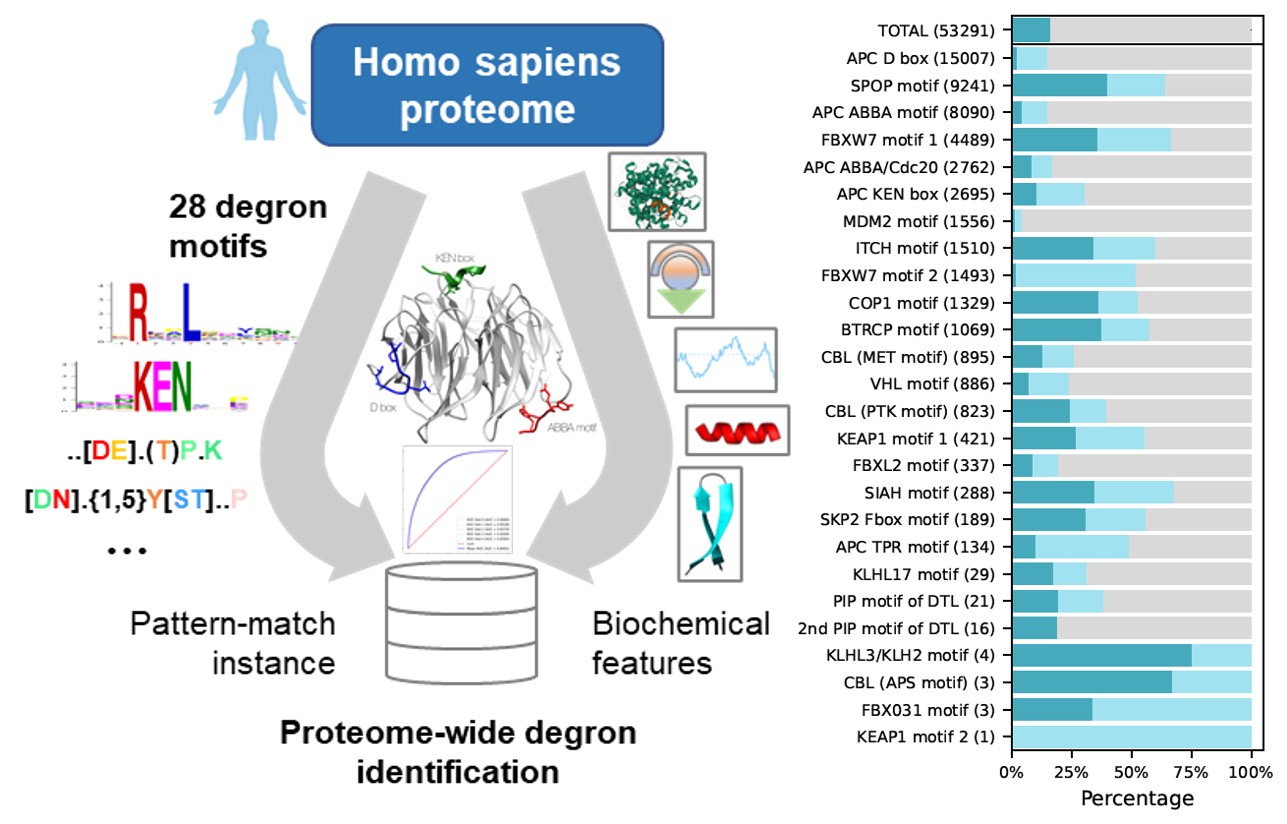
Mar 07, 2022: Scanning the human proteome (Over 20,000 reviewed human proteins downloaded from UniProt) with the 26 known degron motifs produced 53,291 matches.
Feb 02, 2022: Degron motifs i.e., consensus motifs of particular E3 ubiquitin ligase or adaptor protein, in the human were downloaded from ELM database.
Mar 12, 2023: DegronMD website and the main functions include, browsing, querying, and visualization are implemented.
Feb 10, 2023: We systematically investigated and quantified the impact of missense mutations on the functionality of a degron.
Dec 06, 2022: These mutations were then mapped to the regions of all new degron instances resulting in 288,345 somatic mutation records on 122,503 unique amino acid positions.
Nov 26, 2022: ∼7.2 million somatic mutation records were curated from five cancer-related datasets: CCLE, GDSC, TCGA, GDSC and COSMIC.
Oct 28, 2022: 18,929 new degron instances were idenfication in more than 7000 different protein.
Oct 12, 2022: A XGBoost classifier trained on known degron instances by incorporating consensus motifs with multiple degron-intrinsic structural and physicochemical features.
Mar 07, 2022: Scanning the human proteome (Over 20,000 reviewed human proteins downloaded from UniProt) with the 26 known degron motifs produced 53,291 matches.
Feb 02, 2022: Degron motifs i.e., consensus motifs of particular E3 ubiquitin ligase or adaptor protein, in the human were downloaded from ELM database.
Developers:
Haodong Xu, Rufieng Hu, Zhongming Zhao @ CPH, SBMI, UTHealth-Houston.
Citation of the Database:
Haodong Xu, Ruifeng Hu, Zhongming Zhao, DegronMD: Degron mutations and drug response database. (Submitted)