|
|
||
|
|
||
Tutorial for CNVannotator: |
|---|
|
1. Data source of CNVannotator Text search of CNV using gene information Batch search a list of genes in CNVannotator 3. Region-based CNV annotation Overlap with reported common and disease CNVs Annotate with disease information Annotate with genomic features 4. Retrieve submitted jobs and provide feedback to us |
Data source of CNVannotator |
|
The primary aim of CNVannotator is to provide a high-quality CNV annotation portal. CNVannotator, which takes an input set of genomic positions in a user-friendly tabular format that lists the chromosome name with its starting and ending coordinates. In our current release, the genomic coordinates are based on hg19. Please use UCSC LifeOver to convert hg18 coordinates to the hg19 coordinates if necessary. CNVannotator can perform genomic overlaps of the input coordinates with various functional features, including reported 356,817 common CNVs, 181,261 disease CNVs, 140,342 SNPs from genome-wide association studies, protein-coding and non-coding genes and their targets. In addition, CNVannotator is able to incorporate 308,760 genomic features, including cytoband, segmental duplication, pseudogene, promoter, enhancer, CpG island and methylation site. For cancer research community users, CNVannotator can apply various filters to retrieve a subgroup of CNVs according to hundreds of tumor suppressors and oncogenes. In total, 1,044,527 genomic coordinates with functional or genomic features are available to generate an output in a plain text format that is free to download. The annnotation for CNVs involves four main types: reported common and disease CNVs, known disease related mutations, various genomic features in the human genome, and cancer-specific genes and mutations. |
Copy number variations |
|
To annotate CNVs, the first step is to survey previously reported CNVs. Users can either filter out these reported CNVs or use the reported CNVs to confirm their discovery. For these aims, we integrate the known common CNVs from latest DGVdb database (Release in 2012-11-23) and disease related CNVs from CNVD database. |
Disease features |
Similar to SNVs, CNVs do not necessarily have a negative effect on human health. However, among the large number of CNVs, some might have an association with, or be directly involved in, diseases and phenotypes such as cancer and neuropsychiatric disorders. To provide an overview of reported disease sites for the interesting CNVs, CNVannotator integrates disease genes from the genetic association database , GWAS catalog, and GWASdb. In addition, to assist cancer-specific investigations, we also include mutated gene sites from the COSMIC database, TSGene, Tumor Associated Gene (TAG) database, and cancer gene census database. |
Genomic features |
Overlapping CNVs to the genome will help users to study the potential genomic mechanism for the interesting CNVs. To achieve this goal, eight genomic features were collected from UCSC genome browser, including segmental duplication regions, breakpoints for chromosomal events, known cytobands, CpG islands, disease methylation sites, potential transcription factor binding sites, gene promoter regions, and reported enhancer regions. |
|
|
All the CNVs and their annotations in our database are searchable. The gene-based text search provides quick access for the data stored in our web server. Text search of CNV using gene informationUsers can search against the CNVannotator by typing the gene symbols. For one interesting gene, use could just input in the seach form
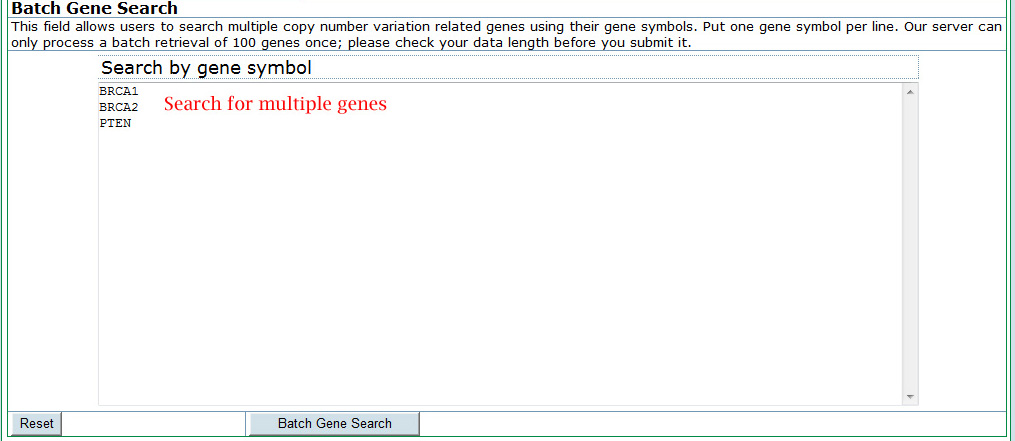
Batch search a list of genes in CNVannotator:To access multiple interesting genes in the database, a batch search form is provided. In the batch search, users can input a list of human gene symbols from the NCBI Entrez database. The exact match with input gene symbols will be sent to find overlapped CNVs, which will be listed in the results page. In the input page, users are required to input their gene symbols line by line.
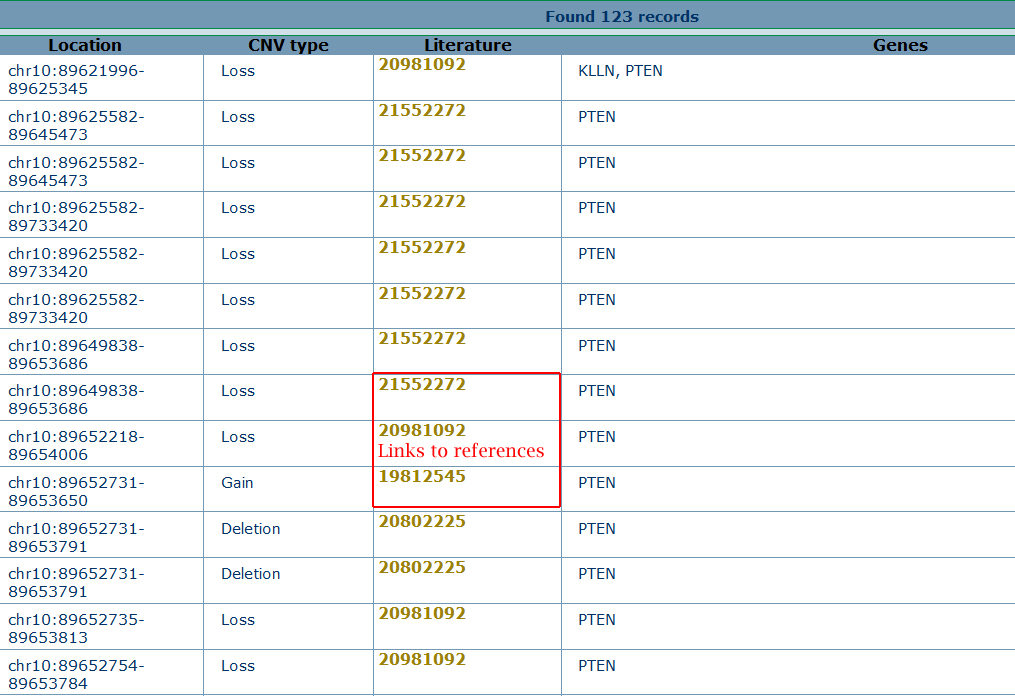
The search results show a list of overlapped CNVs linked as shown in the below.
Click on the hyperlink in the results page, user can access the original references for the reported CNVs. |
Region-based CNV annotation [ top ] |
|
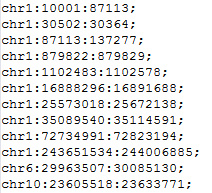
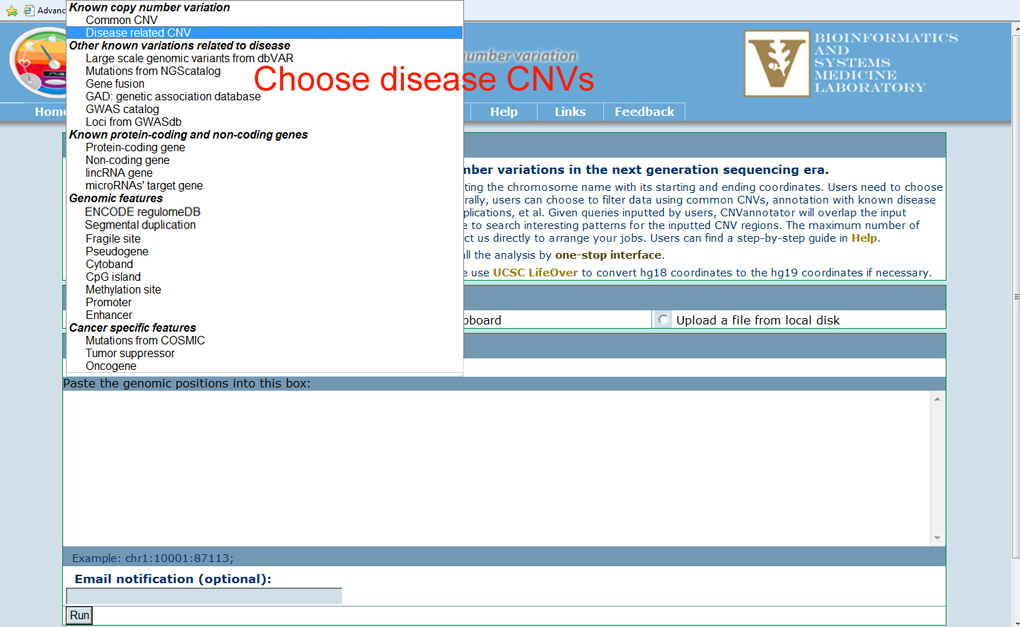
The CNVannotator database supports a search of CNVs using interesting genomic regions. In the region-based search page, users can explore the CNVs with various disease and genomic features. In addition, to help users get a bird's eye view for all the biological information related to the overlapped CNVs, the one-stop annotation search is also provided. First, users can use our example input to take a quick look at the output format. A small number of genomic regions can also be pasted into the text box to search the overlapped CNVs. Finally, users can upload a local file that includes less than 500 genomic regions. The tabular format with chromosome name with its starting and ending coordinates per line is sufficient to begin the CNV annotation. The example input is as the figure below.
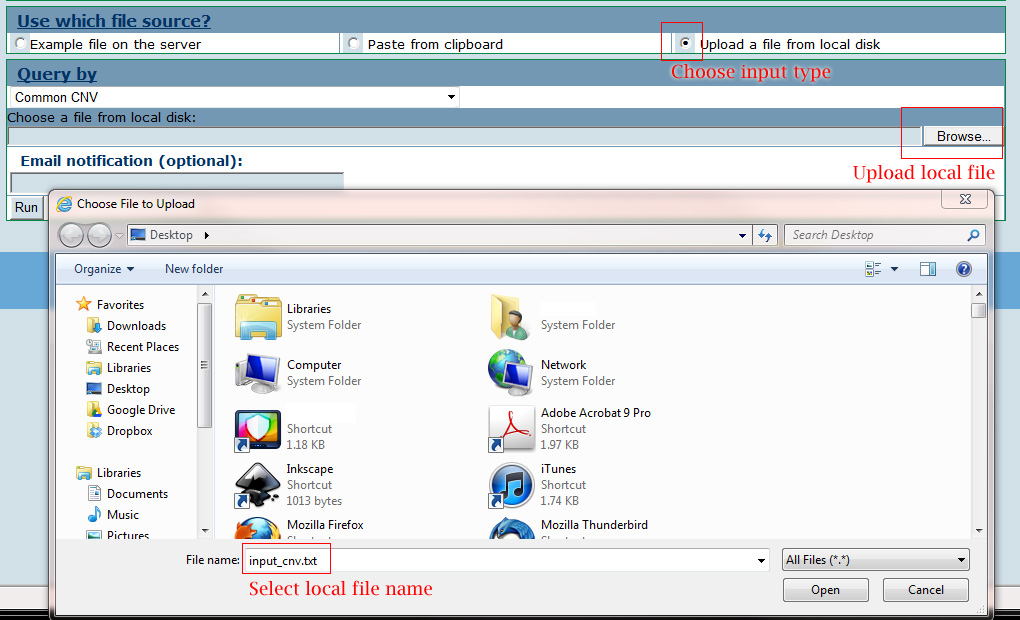
Users need to specify the type of analyses that will be conducted using the drop-down menu. Generally, users can choose to filter data using common CNVs, annotation with known disease CNVs, overlaps with known genes, published GWA studies, segmental duplications, et al. Optionally, users can provide their email address, which is useful for CNVannotator to notify users when the job is finished. Normally, a search is performed within several seconds to minutes. In summary, three basic steps in the CNV annotation page can lead users to obtain their annotation. Take the disease CNV annotation using local file upload as an example below: Input your interesting genomic regions.
Select diesease CNV from from the drop-down menu.
Fill the optional email and then submit the job.
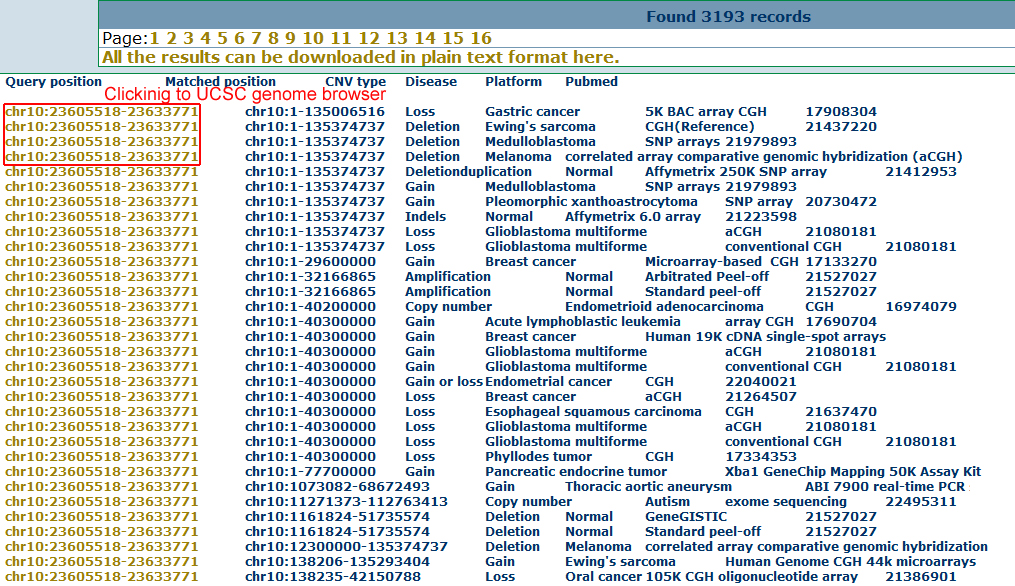
The result shows the list of overlapped CNVs with hyperlinks to the UCSC genome browser.
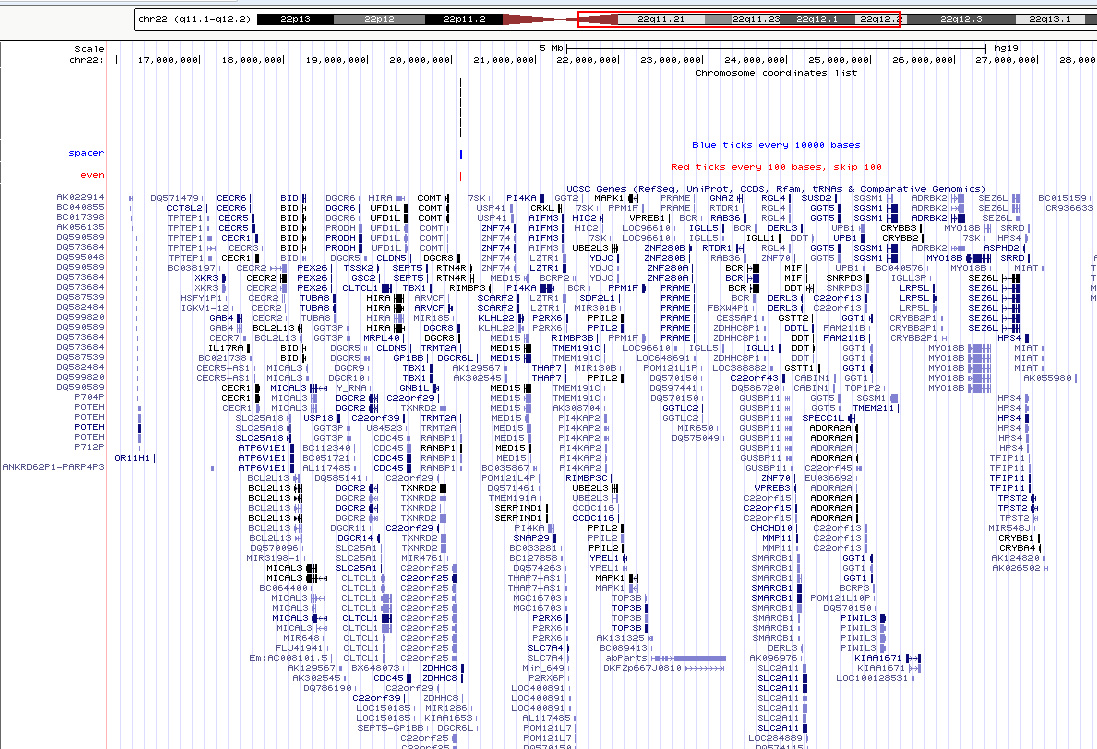
Following the links to UCSC genome browser, user can get more detail information for the interesting genomic regions as below.
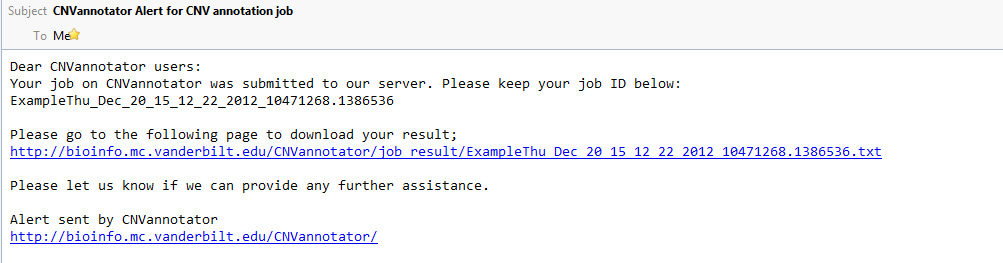
Meanwhile, the reminder email will be sent to users who have filled their email in the email form.
To get a bird's eye view for all the biological information in our CNVannotator, users can use the one-stop annotation to obtain all results as below.
|
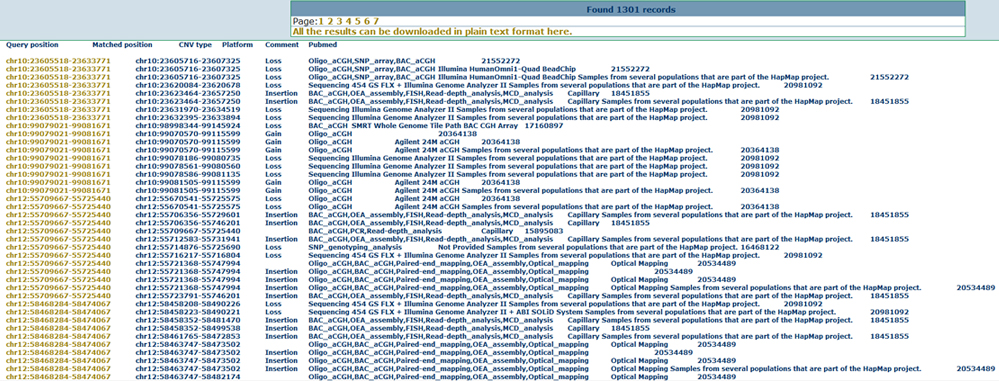
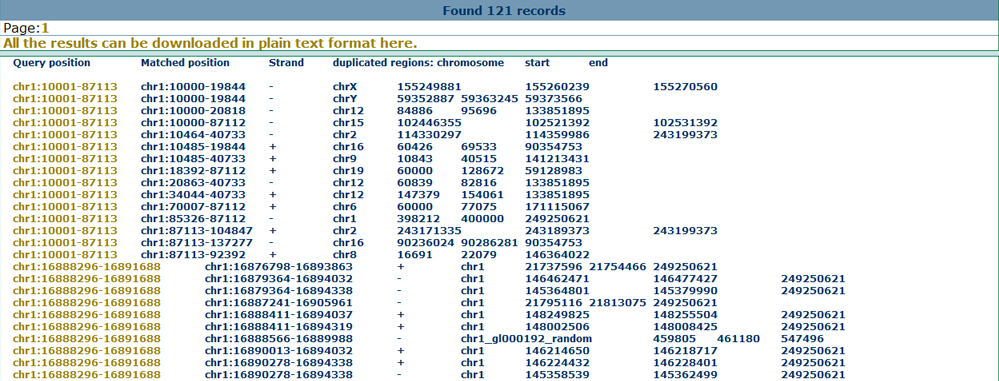
Overlap with reported common and disease CNVsFrom the drop-down menu in the region-based search page, users can obtain the reported common and disease CNV lists from different data sources. This may help users to filter the common CNVs or find causal CNVs in specific disease samples. Here we show the annotation results for the common CNVs as below.
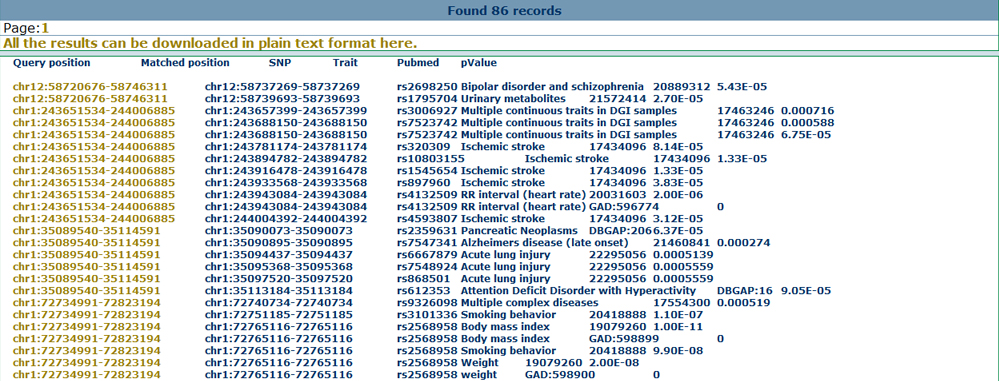
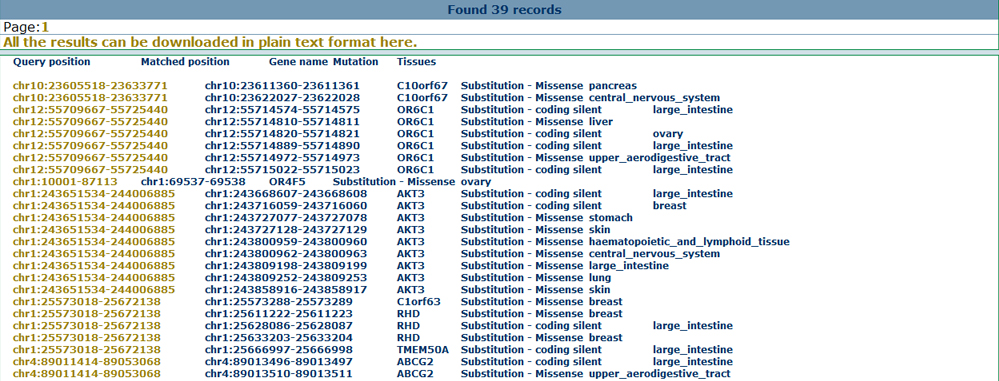
Annotate with disease informationBy mapping the inputted genomic coordinates to disease sites, users can also find all the reported disease related gene or SNP information collected from genomewide studies. Here, we provide the annotation from the genetic association database, GWAS catalog and GWASdb, which may help users to get an overview for the inputted genomic regions on the disease angle. Here we show the annotation results for the significant SNPs from GWASdb as below.
Annotate with genomic featuresThe CNVs are reported as more likely to occur in tandem duplication regions in the human genome. To provide a genomic view of the CNVs, users can obtain 8 different types of genomic features in our CNVannotator server. Here we show the annotation results for the segmental duplicaitons as below.
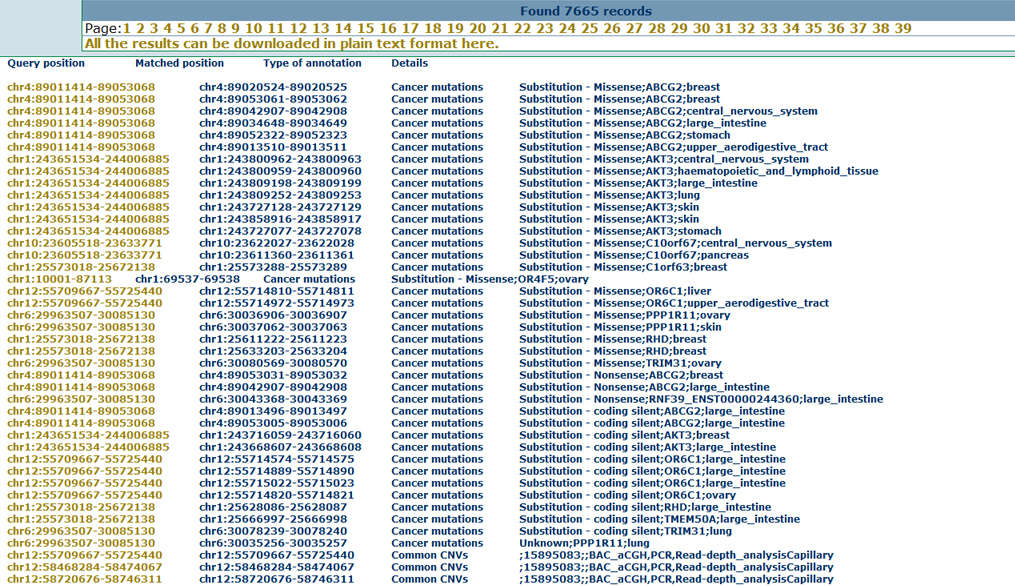
Cancer-specific featuresTo provide a cancer-specific view of the inputted CNVs, users can not only obtain the mutations for various cancer types but also overlap the CNV regions to the important cancer genes, including tumor suppressor genes and oncogenes. Here we show the annotation results for the cancer related mutations as below.
|
Retrieve the submitted jobs and feedback to us [ top ] |
Users can freely download their searched results in plain text format for academic activities but not for profit purposes. Please access the job retrieve page. In the page, user is required to provide the job ID, which has been sent to the user by email or via notification in the results page after submitting job.We also hope you can help us to improve our database.If you have any suggestions or comments regarding the records in the current version of CNVannotator, or to revise incorrect information, please send us this information via feedback page. |