| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 10553 |
Name | HTATIP2 |
Synonymous | HIV-1 Tat interactive protein 2, 30kDa;HTATIP2;HIV-1 Tat interactive protein 2, 30kDa |
Definition | 30 kDa HIV-1 TAT-interacting protein|HIV-1 TAT-interactive protein 2|Tat-interacting protein (30kD)|oxidoreductase HTATIP2|short chain dehydrogenase/reductase family 44U, member 1 |
Position | 11p15.1 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
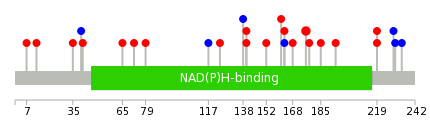 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
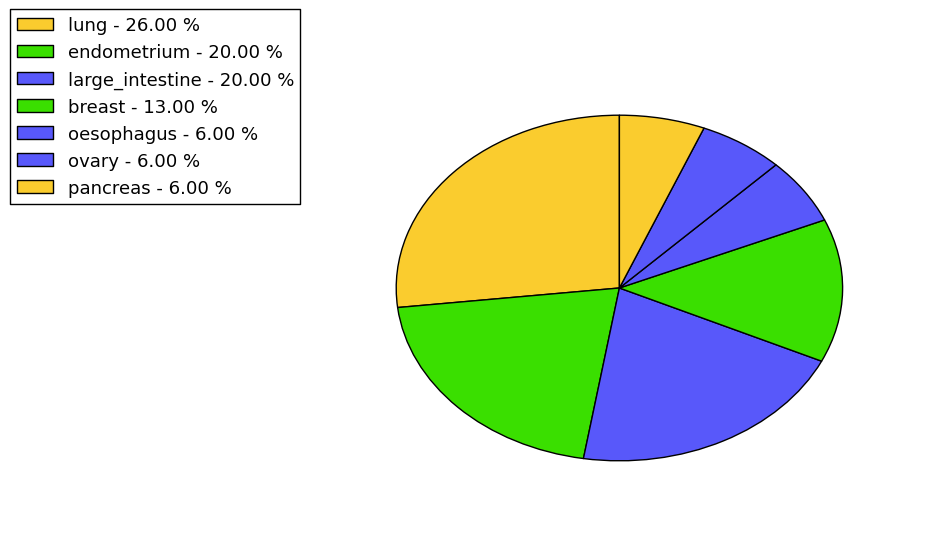 |
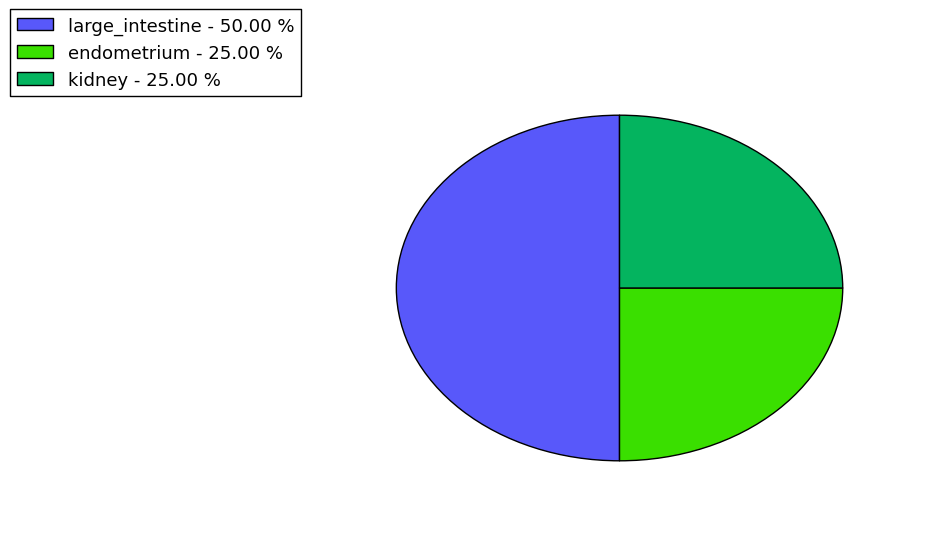 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.13. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.690C>A; p.I230I; 11:20383166-20383166 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.419A>G; p.N140S; 11:20376695-20376695 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.234C>T; p.Y78Y; 11:20367212-20367212 |
bone | Ewings_sarcoma-peripheral_primitive_neuroectodermal_tumour | Substitution - coding silent |
c.526G>C; p.E176Q; 11:20383002-20383002 |
urinary_tract; bladder | carcinoma; transitional_cell_carcinoma | Substitution - Missense |
c.526G>C; p.E176Q; 11:20383002-20383002 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.533G>A; p.R178H; 11:20383009-20383009 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.687C>T; p.A229A; 11:20383163-20383163 |
skin | malignant_melanoma | Substitution - coding silent |
c.489T>C; p.S163S; 11:20382225-20382225 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - coding silent |
c.466G>T; p.E156*; 11:20382202-20382202 |
stomach | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.414A>C; p.S138S; 11:20376690-20376690 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.702G>A; p.G234G; 11:20383178-20383178 |
thyroid | carcinoma | Substitution - coding silent |
c.488C>G; p.S163C; 11:20382224-20382224 |
upper_aerodigestive_tract; pharynx | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.503G>C; p.G168A; 11:20382239-20382239 |
oesophagus | carcinoma; adenocarcinoma | Substitution - Missense |
c.123G>C; p.Q41H; 11:20364360-20364360 |
stomach | adenocarcinoma | Substitution - Missense |
c.553A>G; p.R185G; 11:20383029-20383029 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.37G>A; p.D13N; 11:20364274-20364274 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.376C>T; p.H126Y; 11:20376652-20376652 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.616G>A; p.V206M; 11:20383092-20383092 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.655G>A; p.D219N; 11:20383131-20383131 |
upper_aerodigestive_tract; mouth | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.193G>T; p.V65L; 11:20364430-20364430 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.481C>T; p.R161C; 11:20382217-20382217 |
skin | malignant_melanoma | Substitution - Missense |
c.214T>A; p.F72I; 11:20367192-20367192 |
pancreas | carcinoma | Substitution - Missense |
c.19C>A; p.L7M; 11:20364256-20364256 |
ovary | carcinoma; serous_carcinoma | Substitution - Missense |
c.454G>A; p.A152T; 11:20382190-20382190 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.304-2A>G; p.?; 11:20376578-20376578 |
stomach | carcinoma; adenocarcinoma | Unknown |
c.202G>T; p.E68*; 11:20367180-20367180 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.235G>A; p.A79T; 11:20367213-20367213 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.520_524delAGGCA; p.Q175fs*5; 11:20382996-20383000 |
large_intestine | carcinoma; adenocarcinoma | Deletion - Frameshift |
c.520_524delAGGCA; p.Q175fs*5; 11:20382996-20383000 |
large_intestine | carcinoma; adenocarcinoma | Deletion - Frameshift |
c.656A>G; p.D219G; 11:20383132-20383132 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.419A>C; p.N140T; 11:20376695-20376695 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.120G>A; p.E40E; 11:20364357-20364357 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.581C>T; p.S194F; 11:20383057-20383057 |
breast | carcinoma | Substitution - Missense |
c.104T>C; p.L35S; 11:20364341-20364341 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.413C>G; p.S138*; 11:20376689-20376689 |
breast | carcinoma | Substitution - Nonsense |
c.371G>A; p.C124Y; 11:20376647-20376647 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.351G>A; p.E117E; 11:20376627-20376627 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |