| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 10633 |
Name | RASL10A |
Synonymous | RAS-like, family 10, member A;RASL10A;RAS-like, family 10, member A |
Definition | ras-like protein RRP22|ras-like protein family member 10A|ras-related protein on chromosome 22 |
Position | 22q12.2 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
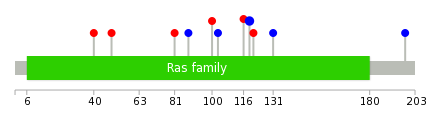 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
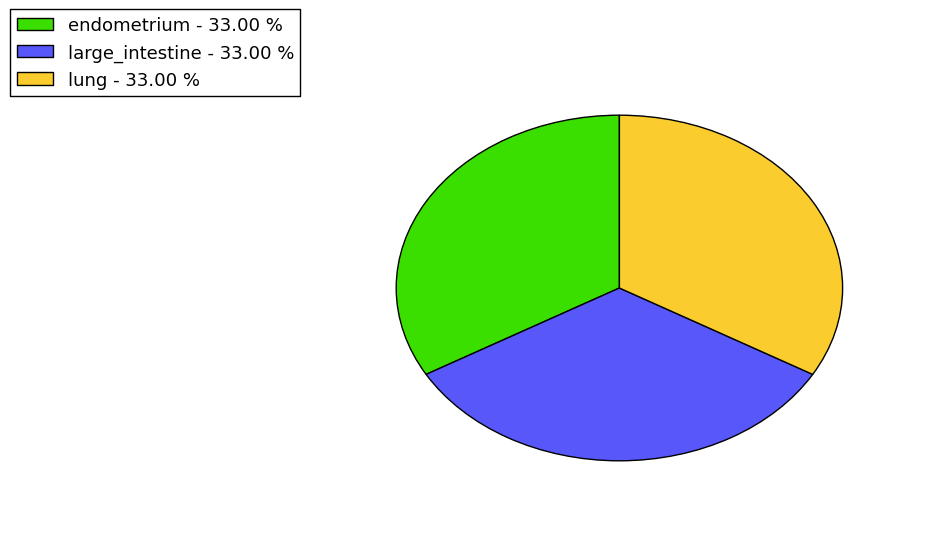 |
 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.00. |
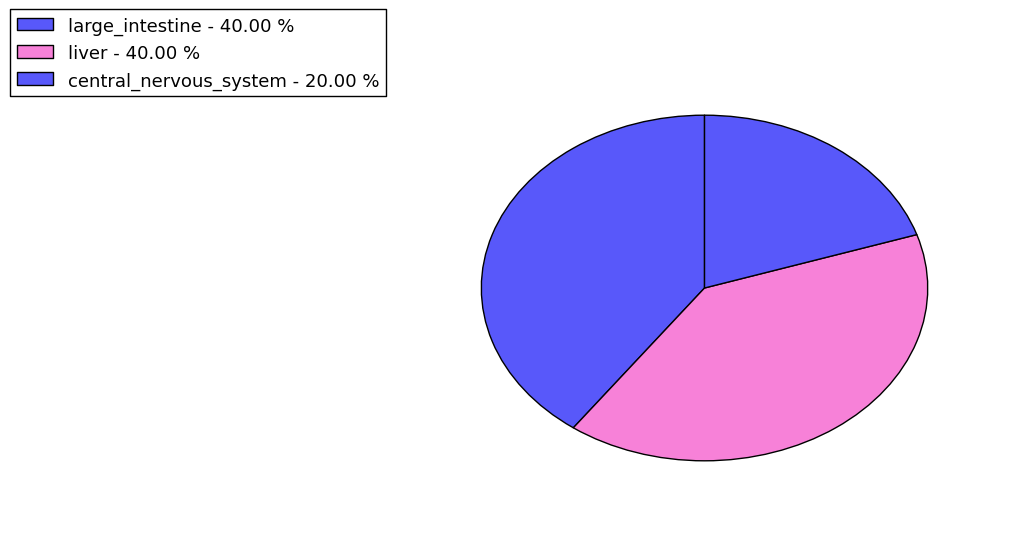
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.347C>T; p.P116L; 22:29313566-29313566 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.594G>A; p.A198A; 22:29313319-29313319 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.393G>A; p.R131R; 22:29313520-29313520 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.361G>A; p.E121K; 22:29313552-29313552 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.357G>A; p.A119A; 22:29313556-29313556 |
liver | carcinoma | Substitution - coding silent |
c.357G>A; p.A119A; 22:29313556-29313556 |
liver | carcinoma | Substitution - coding silent |
c.264C>T; p.A88A; 22:29313943-29313943 |
skin | malignant_melanoma | Substitution - coding silent |
c.309C>T; p.Y103Y; 22:29313898-29313898 |
central_nervous_system; brain | glioma | Substitution - coding silent |
c.243G>C; p.W81C; 22:29313964-29313964 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.299G>T; p.S100I; 22:29313908-29313908 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.119G>C; p.R40P; 22:29315128-29315128 |
NS | NS | Substitution - Missense |
c.428C>T; p.A143V; 22:29313485-29313485 |
oesophagus | carcinoma; adenocarcinoma | Substitution - Missense |
c.445C>T; p.R149C; 22:29313468-29313468 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.331_332insGCGCA; p.I111fs*19; 22:29313875-29313876 |
large_intestine; colon | carcinoma; adenocarcinoma | Insertion - Frameshift |
c.145G>T; p.D49Y; 22:29315102-29315102 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |