| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 148252 |
Name | DIRAS1 |
Synonymous | DIRAS family, GTP-binding RAS-like 1;DIRAS1;DIRAS family, GTP-binding RAS-like 1 |
Definition | GTP-binding protein Di-Ras1|distinct subgroup of the Ras family member 1|ras-related inhibitor of cell growth|small GTP-binding tumor suppressor 1 |
Position | 19p13.3 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
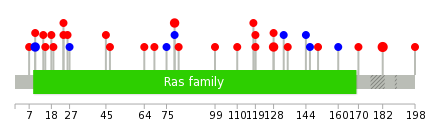 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
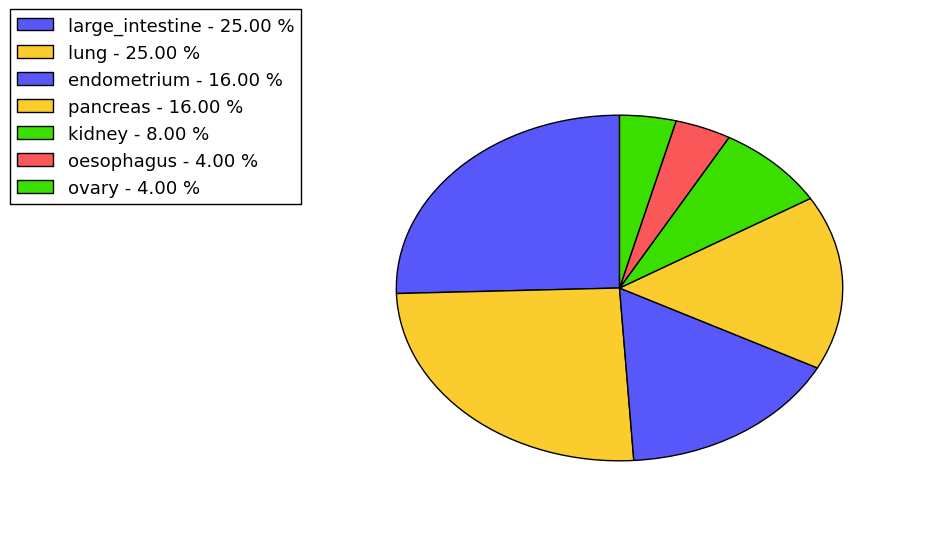 |
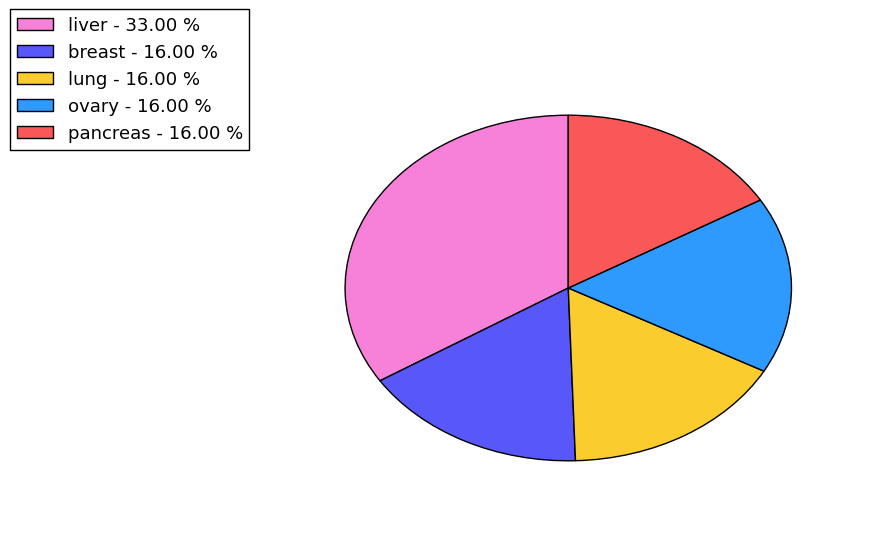 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.04. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.469G>A; p.V157I; 19:2717338-2717338 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.383G>A; p.R128Q; 19:2717424-2717424 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.329G>A; p.S110N; 19:2717478-2717478 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.44C>T; p.A15V; 19:2717763-2717763 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.242C>G; p.A81G; 19:2717565-2717565 |
stomach | adenocarcinoma | Substitution - Missense |
c.70G>T; p.V24L; 19:2717737-2717737 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.406C>T; p.Q136*; 19:2717401-2717401 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.52G>A; p.V18M; 19:2717755-2717755 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.480C>T; p.L160L; 19:2717327-2717327 |
skin | malignant_melanoma | Substitution - coding silent |
c.352C>T; p.L118F; 19:2717455-2717455 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.28G>A; p.V10M; 19:2717779-2717779 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.134G>A; p.R45Q; 19:2717673-2717673 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.592A>G; p.M198V; 19:2717215-2717215 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.237C>T; p.G79G; 19:2717570-2717570 |
ovary | other; neoplasm | Substitution - coding silent |
c.509G>A; p.R170H; 19:2717298-2717298 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.278C>A; p.S93*; 19:2717529-2717529 |
pancreas | carcinoma | Substitution - Nonsense |
c.139G>A; p.V47M; 19:2717668-2717668 |
ovary | carcinoma; serous_carcinoma | Substitution - Missense |
c.27C>T; p.R9R; 19:2717780-2717780 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.345C>T; p.P115P; 19:2717462-2717462 |
oesophagus; lower_third | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.55G>T; p.G19C; 19:2717752-2717752 |
prostate | carcinoma | Substitution - Missense |
c.19G>A; p.D7N; 19:2717788-2717788 |
oesophagus; lower_third | carcinoma; adenocarcinoma | Substitution - Missense |
c.296C>T; p.P99L; 19:2717511-2717511 |
pancreas | carcinoma | Substitution - Missense |
c.87G>T; p.K29N; 19:2717720-2717720 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.225C>T; p.S75S; 19:2717582-2717582 |
pancreas | carcinoma | Substitution - coding silent |
c.355G>A; p.V119M; 19:2717452-2717452 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.235G>T; p.G79C; 19:2717572-2717572 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.235G>T; p.G79C; 19:2717572-2717572 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.432C>T; p.C144C; 19:2717375-2717375 |
lung; left_lower_lobe | carcinoma; adenocarcinoma | Substitution - coding silent |
c.279G>A; p.S93S; 19:2717528-2717528 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.399C>T; p.R133R; 19:2717408-2717408 |
breast | carcinoma | Substitution - coding silent |
c.545C>T; p.S182F; 19:2717262-2717262 |
skin | malignant_melanoma | Substitution - Missense |
c.545C>T; p.S182F; 19:2717262-2717262 |
skin | malignant_melanoma | Substitution - Missense |
c.545C>T; p.S182F; 19:2717262-2717262 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.404C>T; p.A135V; 19:2717403-2717403 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.438C>T; p.F146F; 19:2717369-2717369 |
skin | malignant_melanoma | Substitution - coding silent |
c.41G>T; p.G14V; 19:2717766-2717766 |
prostate | carcinoma; adenocarcinoma | Substitution - Missense |
c.356T>A; p.V119E; 19:2717451-2717451 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.190G>T; p.G64C; 19:2717617-2717617 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.547G>A; p.G183R; 19:2717260-2717260 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.382C>T; p.R128W; 19:2717425-2717425 |
pancreas | carcinoma; ductal_carcinoma | Substitution - Missense |
c.382C>T; p.R128W; 19:2717425-2717425 |
pancreas | carcinoma | Substitution - Missense |
c.30G>T; p.V10V; 19:2717777-2717777 |
liver | carcinoma | Substitution - coding silent |
c.30G>T; p.V10V; 19:2717777-2717777 |
liver | carcinoma | Substitution - coding silent |
c.206C>T; p.P69L; 19:2717601-2717601 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.5C>T; p.P2L; 19:2717802-2717802 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.71T>A; p.V24E; 19:2717736-2717736 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.136C>T; p.Q46*; 19:2717671-2717671 |
skin | malignant_melanoma | Substitution - Nonsense |
c.314T>C; p.V105A; 19:2717493-2717493 |
stomach | carcinoma; intestinal_adenocarcinoma | Substitution - Missense |
c.77G>A; p.R26H; 19:2717730-2717730 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.81C>T; p.F27F; 19:2717726-2717726 |
skin | malignant_melanoma | Substitution - coding silent |
c.449C>T; p.S150L; 19:2717358-2717358 |
skin | malignant_melanoma | Substitution - Missense |