| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 27129 |
Name | HSPB7 |
Synonymous | heat shock 27kDa protein family, member 7 (cardiovascular);HSPB7;heat shock 27kDa protein family, member 7 (cardiovascular) |
Definition | cardiovascular heat shock protein|heat shock 27kD protein family, member 7 (cardiovascular)|heat shock protein beta-7 |
Position | 1p36.13 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
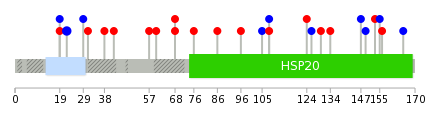 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
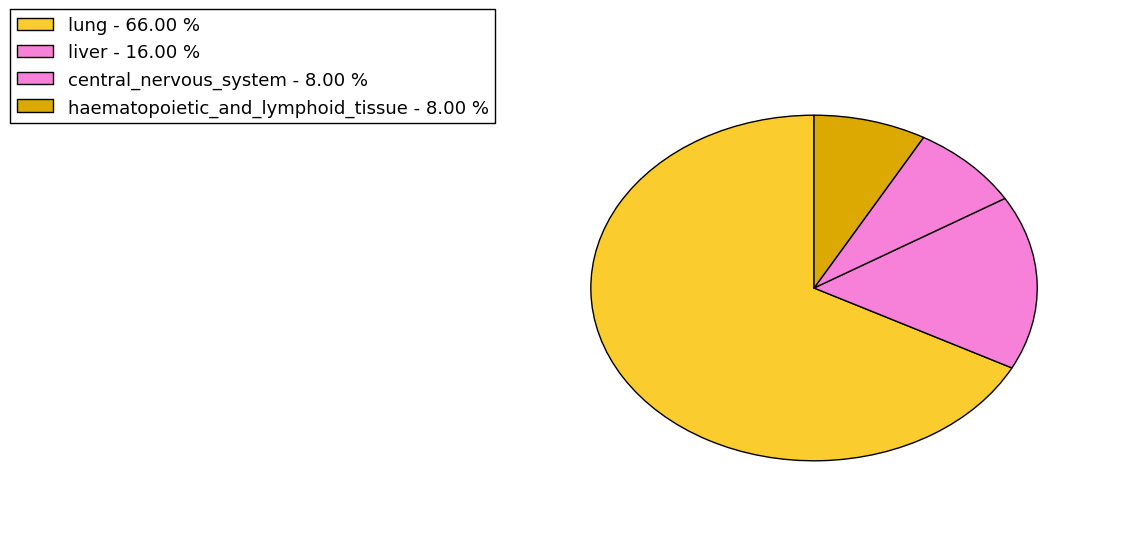 |
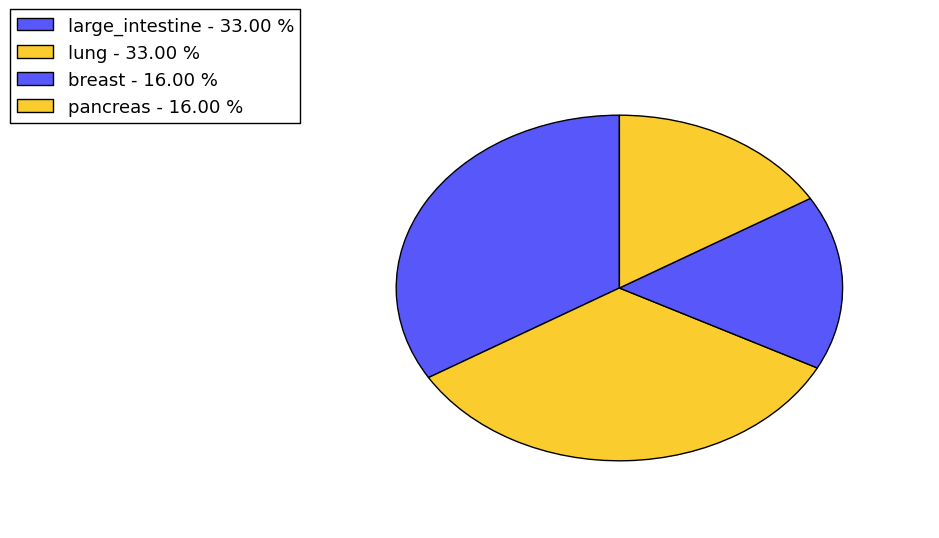 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.23. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.179C>A; p.S60*; 1:16017785-16017785 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Nonsense |
c.441C>A; p.L147L; 1:16015652-16015652 |
thyroid | other; neoplasm | Substitution - coding silent |
c.360C>T; p.N120N; 1:16015733-16015733 |
bone; femur | Ewings_sarcoma-peripheral_primitive_neuroectodermal_tumour | Substitution - coding silent |
c.56C>A; p.S19Y; 1:16017908-16017908 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.124G>A; p.E42K; 1:16017840-16017840 |
liver | carcinoma | Substitution - Missense |
c.254C>T; p.A85V; 1:16017153-16017153 |
skin | malignant_melanoma | Substitution - Missense |
c.466C>G; p.H156D; 1:16015627-16015627 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.399C>T; p.D133D; 1:16015694-16015694 |
bone | Ewings_sarcoma-peripheral_primitive_neuroectodermal_tumour | Substitution - coding silent |
c.145delT; p.S49fs*30; 1:16017819-16017819 |
large_intestine; caecum | carcinoma; adenocarcinoma | Deletion - Frameshift |
c.112G>A; p.D38N; 1:16017852-16017852 |
skin | malignant_melanoma | Substitution - Missense |
c.512G>A; p.*171*; 1:16015581-16015581 |
breast | carcinoma | Substitution - coding silent |
c.458G>A; p.R153H; 1:16015635-16015635 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - Missense |
c.169C>T; p.R57W; 1:16017795-16017795 |
liver | carcinoma; hepatocellular_carcinoma | Substitution - Missense |
c.92C>T; p.S31F; 1:16017872-16017872 |
skin; trunk | malignant_melanoma | Substitution - Missense |
c.316G>T; p.E106*; 1:16017091-16017091 |
lung | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.465G>T; p.P155P; 1:16015628-16015628 |
lung | carcinoma; adenocarcinoma | Substitution - coding silent |
c.203G>A; p.R68H; 1:16017204-16017204 |
central_nervous_system; brain | glioma | Substitution - Missense |
c.207C>T; p.P69P; 1:16017200-16017200 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - coding silent |
c.202C>T; p.R68C; 1:16017205-16017205 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.228G>C; p.K76N; 1:16017179-16017179 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.286A>T; p.I96F; 1:16017121-16017121 |
lung | carcinoma; small_cell_carcinoma | Substitution - Missense |
c.66C>A; p.S22S; 1:16017898-16017898 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.447C>T; p.I149I; 1:16015646-16015646 |
skin | malignant_melanoma | Substitution - coding silent |
c.179C>G; p.S60W; 1:16017785-16017785 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.66C>T; p.S22S; 1:16017898-16017898 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.322C>A; p.R108R; 1:16017085-16017085 |
lung | carcinoma; adenocarcinoma | Substitution - coding silent |
c.495G>A; p.R165R; 1:16015598-16015598 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.378C>T; p.C126C; 1:16015715-16015715 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.388G>A; p.E130K; 1:16015705-16015705 |
skin | malignant_melanoma | Substitution - Missense |
c.370C>G; p.H124D; 1:16015723-16015723 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.257T>A; p.V86E; 1:16017150-16017150 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.87G>A; p.S29S; 1:16017877-16017877 |
pancreas | carcinoma | Substitution - coding silent |
c.339G>A; p.A113A; 1:16015754-16015754 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.57C>T; p.S19S; 1:16017907-16017907 |
thyroid | other; neoplasm | Substitution - coding silent |
c.123G>A; p.M41I; 1:16017841-16017841 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.323G>A; p.R108Q; 1:16017084-16017084 |
skin | malignant_melanoma | Substitution - Missense |
c.400C>T; p.P134S; 1:16015693-16015693 |
skin | malignant_melanoma | Substitution - Missense |
c.315C>T; p.I105I; 1:16017092-16017092 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |