| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 283349 |
Name | RASSF3 |
Synonymous | Ras association (RalGDS/AF-6) domain family member 3;RASSF3;Ras association (RalGDS/AF-6) domain family member 3 |
Definition | Ras association (RalGDS/AF-6) domain family 3|ras association domain-containing protein 3 |
Position | 12q14.2 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
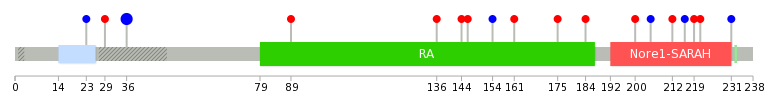 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
 |
 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.00. |
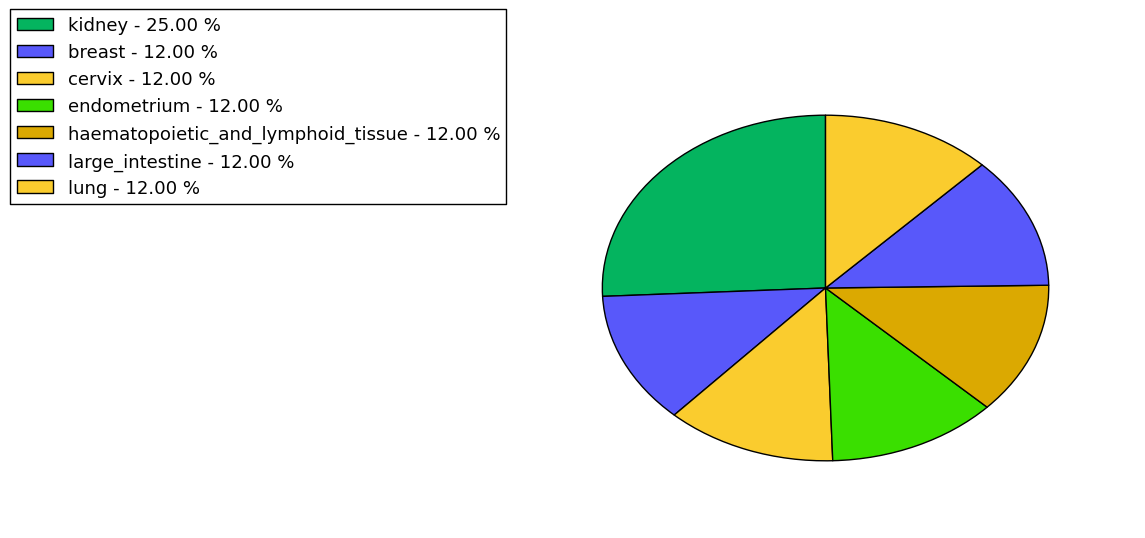
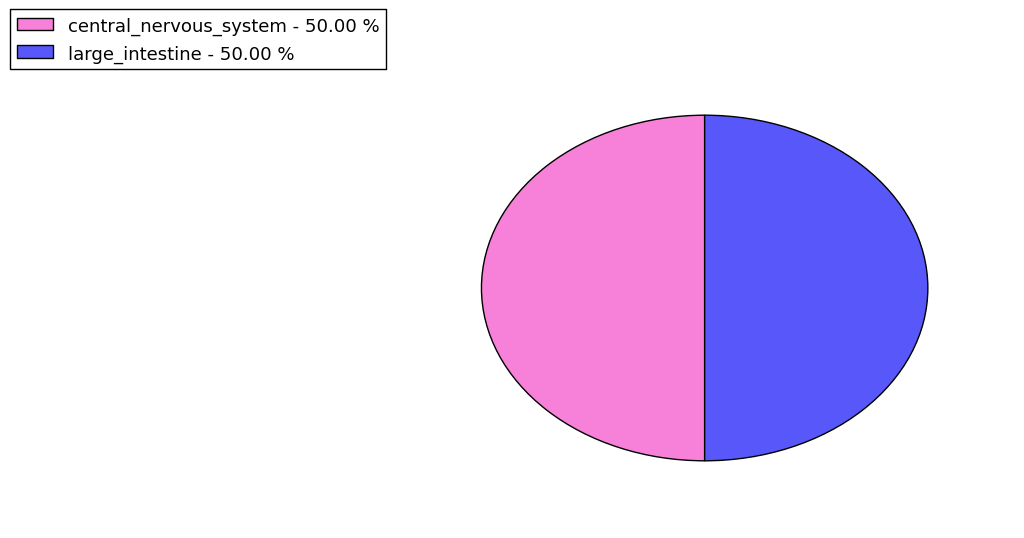
The copy number variations for various cancers | Top |
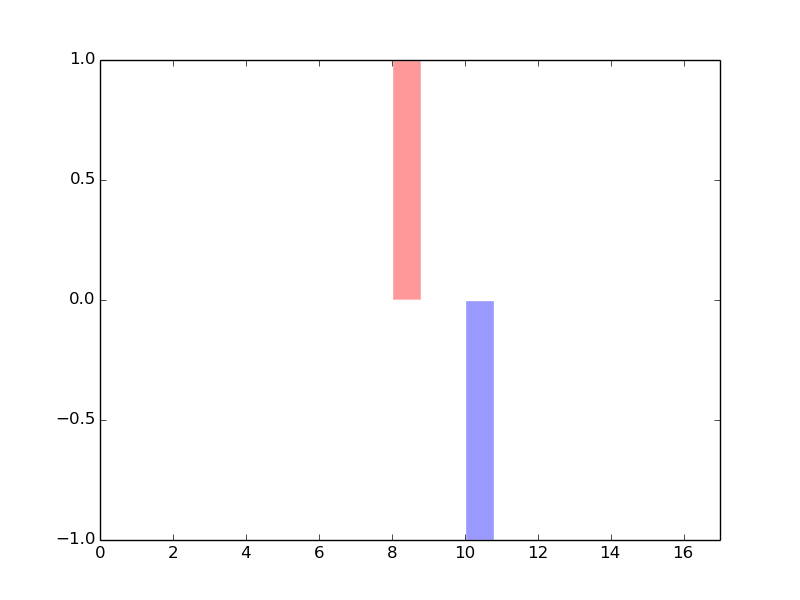 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.523A>G; p.T175A; 12:64691535-64691535 |
breast | carcinoma; basal_(triple-negative)_carcinoma | Substitution - Missense |
c.265C>G; p.L89V; 12:64688261-64688261 |
prostate | carcinoma | Substitution - Missense |
c.693C>T; p.L231L; 12:64694888-64694888 |
upper_aerodigestive_tract; mouth | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.481C>T; p.R161W; 12:64691493-64691493 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.550G>C; p.E184Q; 12:64691562-64691562 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.102C>T; p.S34S; 12:64610734-64610734 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.603C>A; p.F201L; 12:64694798-64694798 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.646C>T; p.L216L; 12:64694841-64694841 |
skin | malignant_melanoma | Substitution - coding silent |
c.598A>G; p.N200D; 12:64694793-64694793 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.108A>G; p.Q36Q; 12:64610740-64610740 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.108A>G; p.Q36Q; 12:64610740-64610740 |
central_nervous_system; brain | glioma; astrocytoma_Grade_IV | Substitution - coding silent |
c.108A>G; p.Q36Q; 12:64610740-64610740 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.108A>G; p.Q36Q; 12:64610740-64610740 |
thyroid | other; neoplasm | Substitution - coding silent |
c.108A>G; p.Q36Q; 12:64610740-64610740 |
central_nervous_system; brain | glioma; astrocytoma_Grade_IV | Substitution - coding silent |
c.108A>G; p.Q36Q; 12:64610740-64610740 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.108A>G; p.Q36Q; 12:64610740-64610740 |
central_nervous_system; brain | glioma; astrocytoma_Grade_IV | Substitution - coding silent |
c.108A>G; p.Q36Q; 12:64610740-64610740 |
central_nervous_system; brain | glioma; astrocytoma_Grade_IV | Substitution - coding silent |
c.69C>T; p.F23F; 12:64610701-64610701 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.431A>G; p.Y144C; 12:64688427-64688427 |
lung | carcinoma; small_cell_carcinoma | Substitution - Missense |
c.662C>A; p.T221K; 12:64694857-64694857 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.613T>C; p.L205L; 12:64694808-64694808 |
skin | malignant_melanoma | Substitution - coding silent |
c.86A>G; p.Q29R; 12:64610718-64610718 |
skin | malignant_melanoma | Substitution - Missense |
c.406G>A; p.E136K; 12:64688402-64688402 |
cervix | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.608G>A; p.R203H; 12:64694803-64694803 |
oesophagus; lower_third | carcinoma; adenocarcinoma | Substitution - Missense |
c.437G>A; p.R146H; 12:64688433-64688433 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - Missense |
c.43_44TT>GA; p.F15>?; 12:64610675-64610676 |
lung | carcinoma; adenocarcinoma | Complex |
c.267C>T; p.L89L; 12:64688263-64688263 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.462C>T; p.Y154Y; 12:64691474-64691474 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.220-9C>T; p.?; 12:64688207-64688207 |
liver | carcinoma | Unknown |
c.601T>G; p.F201V; 12:64694796-64694796 |
stomach | carcinoma; intestinal_adenocarcinoma | Substitution - Missense |
c.72C>T; p.F24F; 12:64610704-64610704 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.42_44delCTT; p.F17delF; 12:64610674-64610676 |
large_intestine; colon | carcinoma; adenocarcinoma | Deletion - In frame |
c.111+2T>C; p.?; 12:64610745-64610745 |
lung | carcinoma; small_cell_carcinoma | Unknown |
c.567+8A>T; p.?; 12:64691587-64691587 |
skin; arm | malignant_melanoma | Unknown |
c.655C>T; p.R219C; 12:64694850-64694850 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.16A>G; p.S6G; 12:64610648-64610648 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.635A>G; p.Q212R; 12:64694830-64694830 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |