| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 286319 |
Name | TUSC1 |
Synonymous | tumor suppressor candidate 1;TUSC1;tumor suppressor candidate 1 |
Definition | tumor suppressor candidate gene 1 protein |
Position | 9p21.2 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
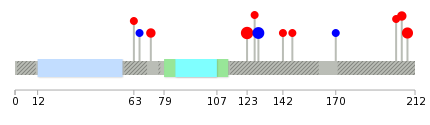 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
 |
 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.00. |
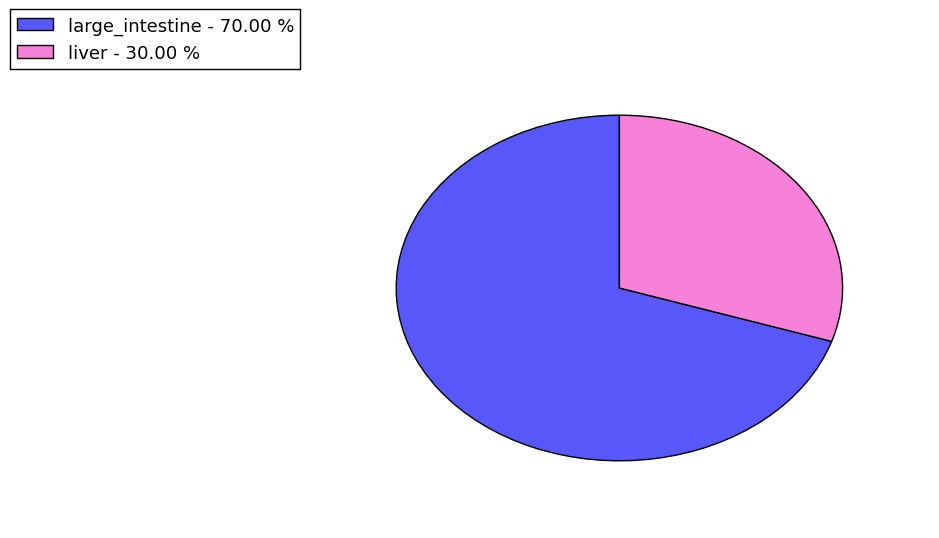
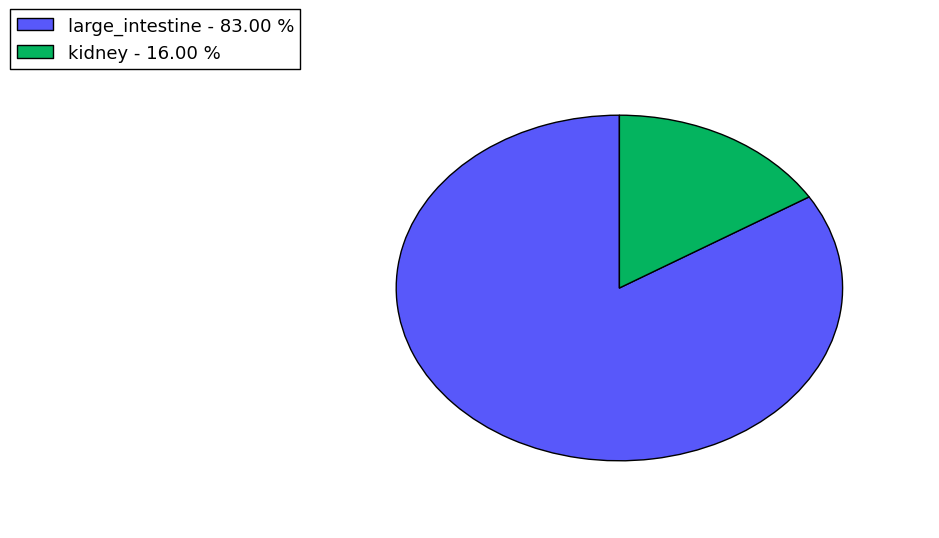
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.424G>T; p.A142S; 9:25677898-25677898 |
liver | carcinoma | Substitution - Missense |
c.614C>T; p.S205L; 9:25677708-25677708 |
liver | carcinoma | Substitution - Missense |
c.614C>T; p.S205L; 9:25677708-25677708 |
liver | carcinoma; hepatocellular_carcinoma | Substitution - Missense |
c.214C>G; p.H72D; 9:25678108-25678108 |
liver | carcinoma | Substitution - Missense |
c.214C>G; p.H72D; 9:25678108-25678108 |
liver | carcinoma | Substitution - Missense |
c.198C>G; p.A66A; 9:25678124-25678124 |
thyroid | other; neoplasm | Substitution - coding silent |
c.367A>G; p.N123D; 9:25677955-25677955 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.367A>G; p.N123D; 9:25677955-25677955 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.367A>G; p.N123D; 9:25677955-25677955 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.367A>G; p.N123D; 9:25677955-25677955 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.367A>G; p.N123D; 9:25677955-25677955 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.367A>G; p.N123D; 9:25677955-25677955 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.367A>G; p.N123D; 9:25677955-25677955 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.367A>G; p.N123D; 9:25677955-25677955 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.367A>G; p.N123D; 9:25677955-25677955 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.142_143insGCG; p.G47_V48insG; 9:25678179-25678180 |
adrenal_gland; adrenal_gland | adrenal_cortical_carcinoma; functioning | Insertion - In frame |
c.509G>A; p.R170Q; 9:25677813-25677813 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.188A>C; p.E63A; 9:25678134-25678134 |
thyroid | carcinoma; medullary_carcinoma | Substitution - Missense |
c.220G>T; p.E74*; 9:25678102-25678102 |
prostate | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.605C>T; p.S202F; 9:25677717-25677717 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.510G>A; p.R170R; 9:25677812-25677812 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - coding silent |
c.380C>T; p.A127V; 9:25677942-25677942 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.440G>T; p.R147L; 9:25677882-25677882 |
liver | carcinoma | Substitution - Missense |
c.387A>G; p.A129A; 9:25677935-25677935 |
thyroid | other; neoplasm | Substitution - coding silent |
c.387A>G; p.A129A; 9:25677935-25677935 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.387A>G; p.A129A; 9:25677935-25677935 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.387A>G; p.A129A; 9:25677935-25677935 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.387A>G; p.A129A; 9:25677935-25677935 |
thyroid | other; neoplasm | Substitution - coding silent |
c.387A>G; p.A129A; 9:25677935-25677935 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.387A>G; p.A129A; 9:25677935-25677935 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.622T>G; p.S208A; 9:25677700-25677700 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.622T>G; p.S208A; 9:25677700-25677700 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.622T>G; p.S208A; 9:25677700-25677700 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.622T>G; p.S208A; 9:25677700-25677700 |
thyroid | other; neoplasm | Substitution - Missense |
c.622T>G; p.S208A; 9:25677700-25677700 |
thyroid | other; neoplasm | Substitution - Missense |
c.335G>A; p.R112H; 9:25677987-25677987 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |