| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 429 |
Name | ASCL1 |
Synonymous | achaete-scute family bHLH transcription factor 1;ASCL1;achaete-scute family bHLH transcription factor 1 |
Definition | ASH-1|achaete scute protein|achaete-scute complex homolog 1|achaete-scute complex-like 1|achaete-scute homolog 1|class A basic helix-loop-helix protein 46 |
Position | 12q23.2 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
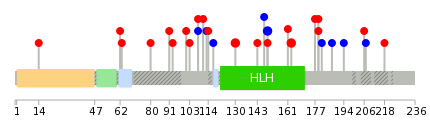 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
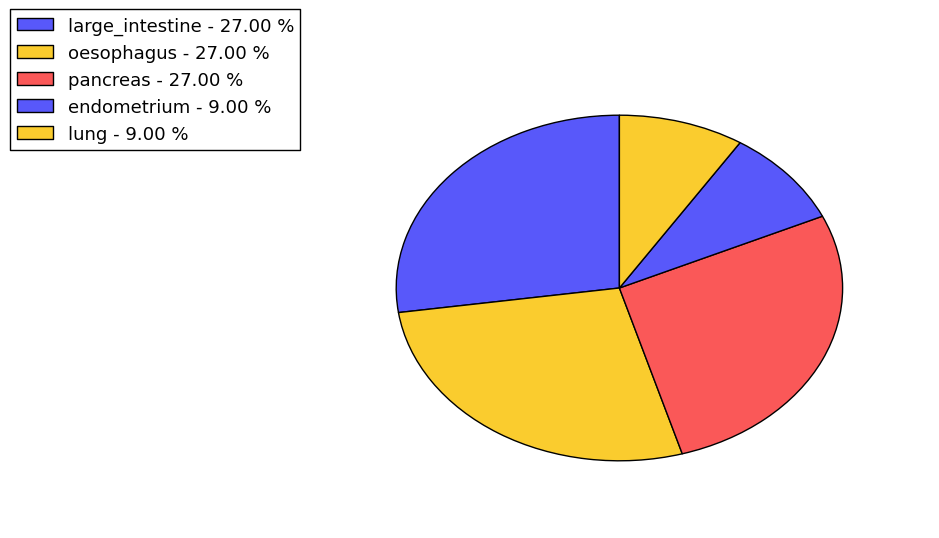 |
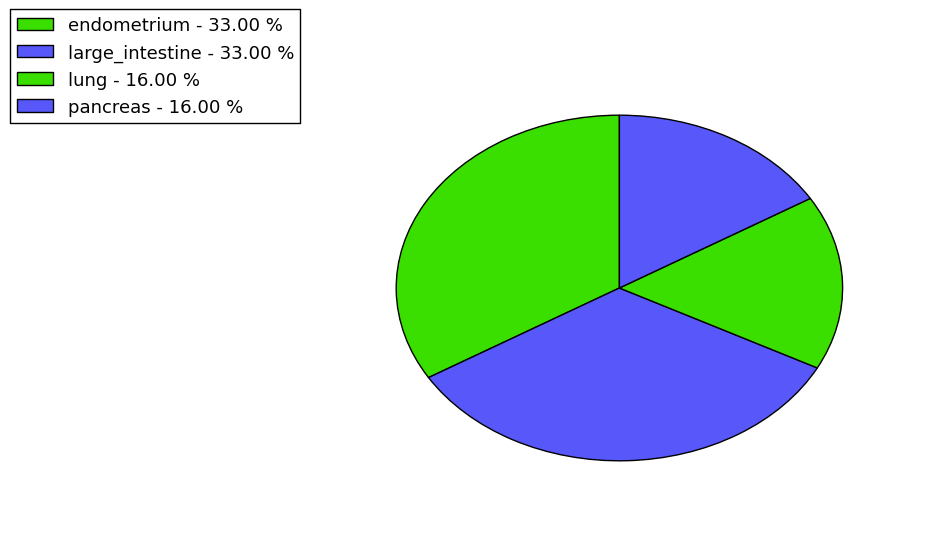 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.17. |
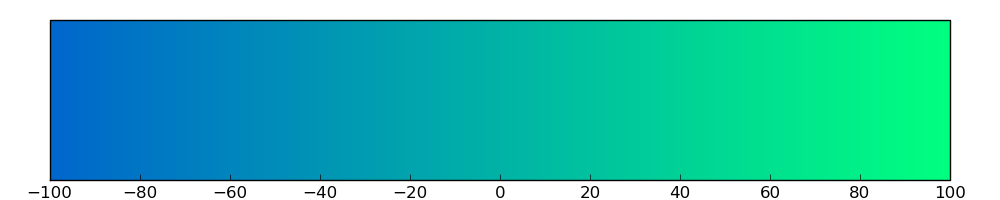
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.389G>A; p.R130H; 12:102958633-102958633 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.341A>T; p.Q114L; 12:102958585-102958585 |
skin | malignant_melanoma | Substitution - Missense |
c.389G>A; p.R130H; 12:102958633-102958633 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.621G>A; p.S207S; 12:102958865-102958865 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.302G>A; p.R101H; 12:102958546-102958546 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.456G>A; p.K152K; 12:102958700-102958700 |
oesophagus; middle_third | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.582C>T; p.S194S; 12:102958826-102958826 |
thyroid | other; neoplasm | Substitution - coding silent |
c.653C>T; p.S218F; 12:102958897-102958897 |
skin | malignant_melanoma | Substitution - Missense |
c.559G>T; p.G187C; 12:102958803-102958803 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.559G>T; p.G187C; 12:102958803-102958803 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.536C>G; p.A179G; 12:102958780-102958780 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.278C>T; p.S93L; 12:102958522-102958522 |
soft_tissue; blood_vessel | angiosarcoma | Substitution - Missense |
c.272C>T; p.S91L; 12:102958516-102958516 |
oesophagus | carcinoma; adenocarcinoma | Substitution - Missense |
c.150_152delGCA; p.Q62delQ; 12:102958394-102958396 |
biliary_tract; bile_duct | carcinoma; adenocarcinoma | Deletion - In frame |
c.471G>A; p.V157V; 12:102958715-102958715 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.383G>A; p.R128H; 12:102958627-102958627 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.149_150insGCA; p.Q62_A63insQ; 12:102958393-102958394 |
large_intestine; colon | carcinoma; adenocarcinoma | Insertion - In frame |
c.149_150insGCA; p.Q62_A63insQ; 12:102958393-102958394 |
large_intestine; colon | carcinoma; adenocarcinoma | Insertion - In frame |
c.149_150insGCA; p.Q62_A63insQ; 12:102958393-102958394 |
large_intestine; colon | carcinoma; adenocarcinoma | Insertion - In frame |
c.561C>G; p.G187G; 12:102958805-102958805 |
lung | carcinoma; adenocarcinoma | Substitution - coding silent |
c.188C>T; p.A63V; 12:102958432-102958432 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.351G>A; p.P117P; 12:102958595-102958595 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.307C>T; p.L103F; 12:102958551-102958551 |
pancreas | carcinoma; acinar_carcinoma | Substitution - Missense |
c.332G>T; p.S111I; 12:102958576-102958576 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.248_249CC>TT; p.P83L; 12:102958492-102958493 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.529C>T; p.H177Y; 12:102958773-102958773 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.239A>G; p.K80R; 12:102958483-102958483 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.41A>T; p.Q14L; 12:102958285-102958285 |
pancreas | carcinoma | Substitution - Missense |
c.616G>A; p.G206S; 12:102958860-102958860 |
oesophagus; lower_third | carcinoma; adenocarcinoma | Substitution - Missense |
c.536C>T; p.A179V; 12:102958780-102958780 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.322T>G; p.F108V; 12:102958566-102958566 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.566T>A; p.L189Q; 12:102958810-102958810 |
stomach | carcinoma; intestinal_adenocarcinoma | Substitution - Missense |
c.429G>C; p.E143D; 12:102958673-102958673 |
pancreas | carcinoma | Substitution - Missense |
c.324T>C; p.F108F; 12:102958568-102958568 |
skin | malignant_melanoma | Substitution - coding silent |
c.481C>A; p.R161S; 12:102958725-102958725 |
thyroid | carcinoma | Substitution - Missense |
c.488_490CGG>CG; p.A163fs; 12:102958732-102958734 |
NS | NS | Complex |
c.488_490CGG>CG; p.A163fs; 12:102958732-102958734 |
NS | NS | Complex |
c.291G>A; p.M97I; 12:102958535-102958535 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.339G>A; p.P113P; 12:102958583-102958583 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.447G>A; p.A149A; 12:102958691-102958691 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.447G>A; p.A149A; 12:102958691-102958691 |
pancreas | carcinoma | Substitution - coding silent |
c.330C>A; p.Y110*; 12:102958574-102958574 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Nonsense |
c.543C>T; p.S181S; 12:102958787-102958787 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.490delG; p.V164fs*17; 12:102958734-102958734 |
NS | NS | Deletion - Frameshift |
c.445G>A; p.A149T; 12:102958689-102958689 |
oesophagus | carcinoma; adenocarcinoma | Substitution - Missense |
c.441C>T; p.N147N; 12:102958685-102958685 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.504C>T; p.R168R; 12:102958748-102958748 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.444C>T; p.G148G; 12:102958688-102958688 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.444C>T; p.G148G; 12:102958688-102958688 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.386A>G; p.N129S; 12:102958630-102958630 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.386A>G; p.N129S; 12:102958630-102958630 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.301C>T; p.R101C; 12:102958545-102958545 |
stomach | carcinoma; diffuse_adenocarcinoma | Substitution - Missense |
c.485C>T; p.S162L; 12:102958729-102958729 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.359T>A; p.V120E; 12:102958603-102958603 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |