| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 4601 |
Name | MXI1 |
Synonymous | MAX interactor 1, dimerization protein;MXI1;MAX interactor 1, dimerization protein |
Definition | MAX dimerization protein 2|Max-related transcription factor|class C basic helix-loop-helix protein 11|max-interacting protein 1 |
Position | 10q24-q25 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
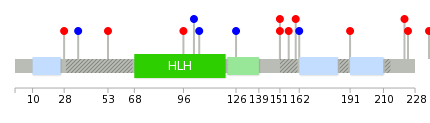 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
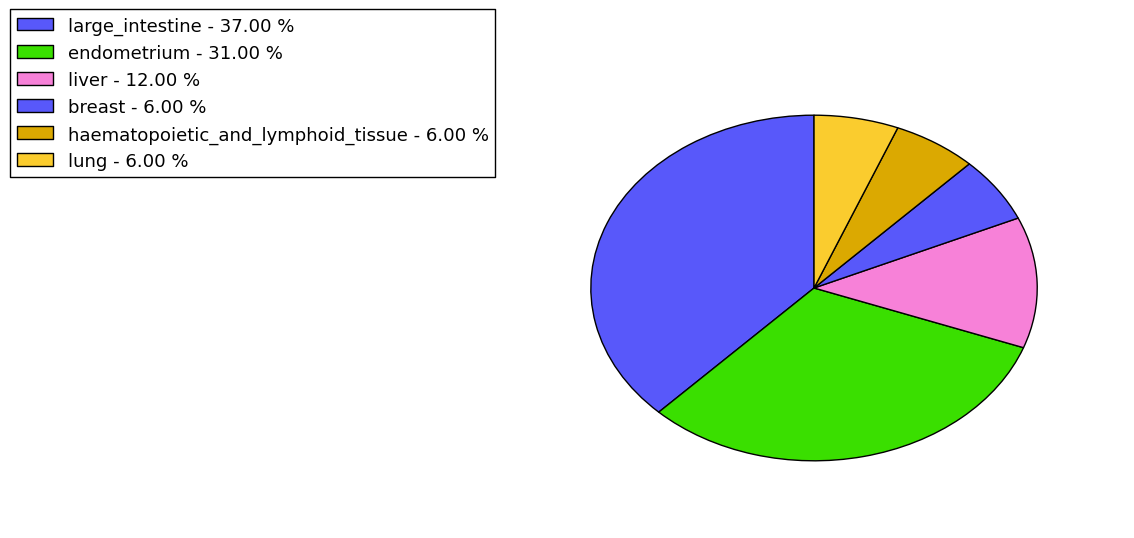 |
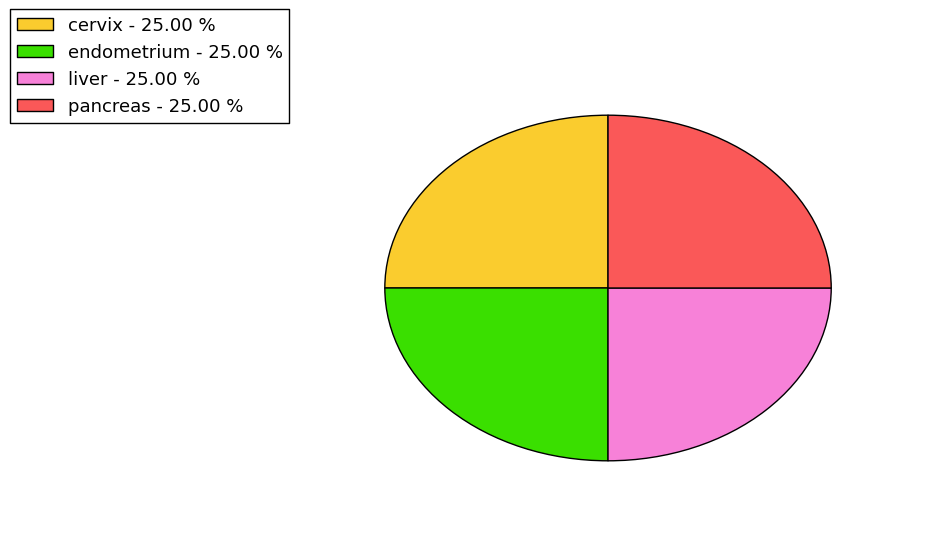 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.24. |
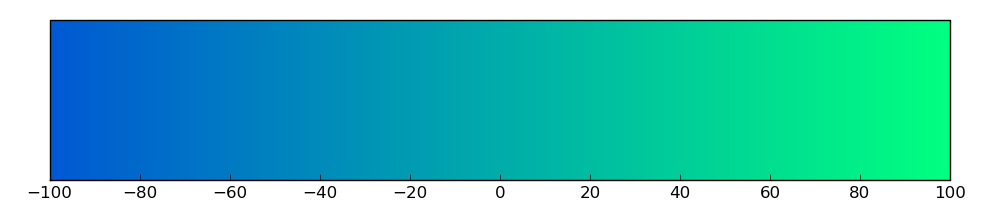
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.831T>C; p.S277S; 10:110284930-110284930 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.812A>G; p.D271G; 10:110284911-110284911 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.479T>C; p.L160P; 10:110279221-110279221 |
thyroid | carcinoma; medullary_carcinoma | Substitution - Missense |
c.707G>A; p.R236H; 10:110280068-110280068 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.671G>A; p.R224Q; 10:110280032-110280032 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.628C>T; p.R210*; 10:110279989-110279989 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.572G>T; p.R191I; 10:110279933-110279933 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.828G>A; p.P276P; 10:110284927-110284927 |
skin | malignant_melanoma | Substitution - coding silent |
c.315G>A; p.P105P; 10:110228229-110228229 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.452G>A; p.R151H; 10:110279194-110279194 |
large_intestine; colon | carcinoma | Substitution - Missense |
c.452G>A; p.R151H; 10:110279194-110279194 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.451C>T; p.R151C; 10:110279193-110279193 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.670C>T; p.R224*; 10:110280031-110280031 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.162T>G; p.I54M; 10:110207970-110207970 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.265G>T; p.E89*; 10:110208073-110208073 |
skin | malignant_melanoma | Substitution - Nonsense |
c.157G>A; p.D53N; 10:110207965-110207965 |
breast | carcinoma; lobular_carcinoma | Substitution - Missense |
c.306G>A; p.P102P; 10:110228220-110228220 |
cervix | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.665G>A; p.R222Q; 10:110280026-110280026 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.108G>A; p.P36P; 10:110207916-110207916 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.781A>G; p.I261V; 10:110284880-110284880 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.725-5G>T; p.?; 10:110284819-110284819 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Unknown |
c.806T>A; p.I269N; 10:110284905-110284905 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.607G>T; p.E203*; 10:110279968-110279968 |
liver | carcinoma | Substitution - Nonsense |
c.607G>T; p.E203*; 10:110279968-110279968 |
liver | carcinoma | Substitution - Nonsense |
c.486A>G; p.P162P; 10:110279228-110279228 |
liver | carcinoma; hepatocellular_carcinoma | Substitution - coding silent |
c.587A>C; p.Q196P; 10:110279948-110279948 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.587A>C; p.Q196P; 10:110279948-110279948 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.844G>A; p.E282K; 10:110284943-110284943 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.266_270delAAAAC; p.N90fs*3; 10:110208074-110208078 |
central_nervous_system; brain | glioma; oligodendroglioma_Grade_III | Deletion - Frameshift |
c.83C>G; p.P28R; 10:110207891-110207891 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; mantle_cell_lymphoma | Substitution - Missense |
c.863G>T; p.S288I; 10:110284962-110284962 |
bone | Ewings_sarcoma-peripheral_primitive_neuroectodermal_tumour | Substitution - Missense |
c.500G>A; p.C167Y; 10:110279242-110279242 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.467G>T; p.R156L; 10:110279209-110279209 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.378C>T; p.S126S; 10:110228292-110228292 |
pancreas | carcinoma | Substitution - coding silent |
c.759C>A; p.F253L; 10:110284858-110284858 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.287G>T; p.G96V; 10:110228201-110228201 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |