| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 56925 |
Name | LXN |
Synonymous | latexin;LXN;latexin |
Definition | MUM|endogenous carboxypeptidase inhibitor|tissue carboxypeptidase inhibitor |
Position | 3q25.32 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
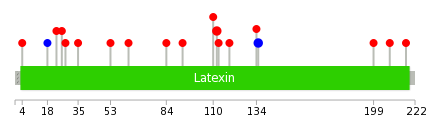 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
 |
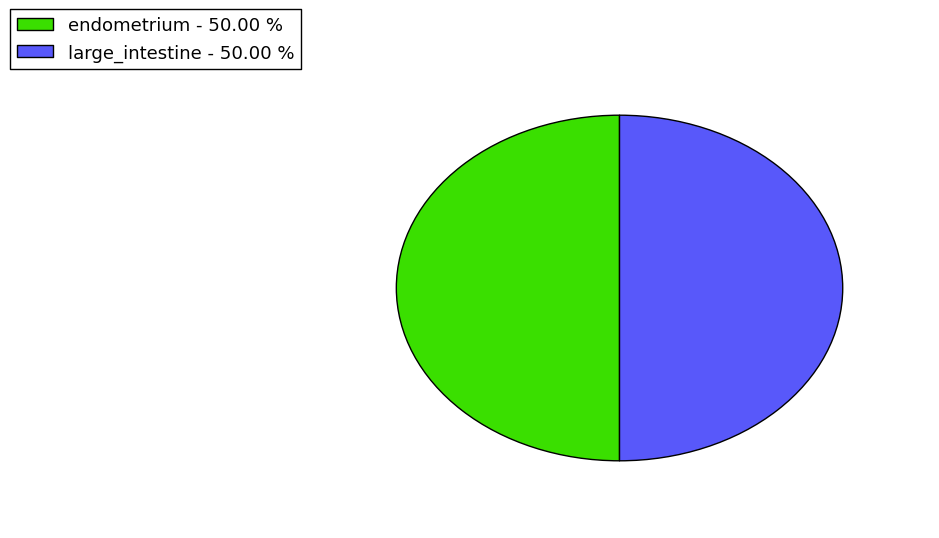 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.00. |
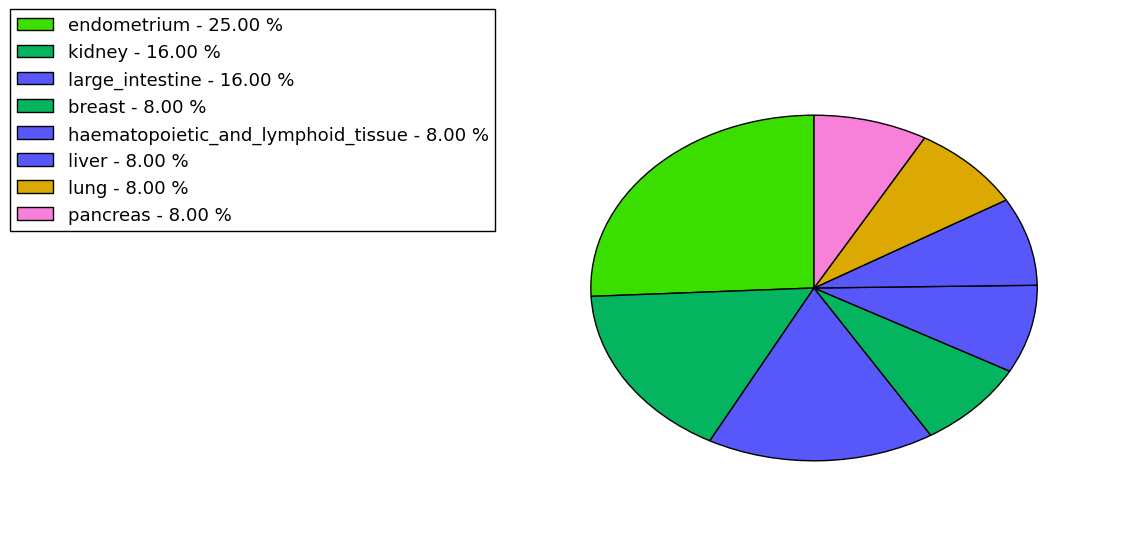
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.158A>G; p.H53R; 3:158670991-158670991 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - Missense |
c.571G>A; p.E191K; 3:158666744-158666744 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.278G>C; p.G93A; 3:158669525-158669525 |
pancreas | carcinoma; acinar_carcinoma | Substitution - Missense |
c.623A>C; p.K208T; 3:158666692-158666692 |
liver | carcinoma | Substitution - Missense |
c.194A>G; p.Q65R; 3:158669609-158669609 |
skin | malignant_melanoma | Substitution - Missense |
c.335C>G; p.S112C; 3:158669468-158669468 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.335C>G; p.S112C; 3:158669468-158669468 |
urinary_tract; bladder | carcinoma; transitional_cell_carcinoma | Substitution - Missense |
c.587A>G; p.Q196R; 3:158666728-158666728 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.189A>C; p.Q63H; 3:158670960-158670960 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.83C>A; p.P28Q; 3:158672396-158672396 |
skin | malignant_melanoma | Substitution - Missense |
c.356C>T; p.A119V; 3:158669447-158669447 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.11C>T; p.P4L; 3:158672468-158672468 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.640C>A; p.R214S; 3:158666675-158666675 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.338T>C; p.M113T; 3:158669465-158669465 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.54G>A; p.Q18Q; 3:158672425-158672425 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.595G>C; p.V199L; 3:158666720-158666720 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.508-1G>A; p.?; 3:158667075-158667075 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Unknown |
c.508-1G>A; p.?; 3:158667075-158667075 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Unknown |
c.508-1G>A; p.?; 3:158667075-158667075 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Unknown |
c.508-1G>A; p.?; 3:158667075-158667075 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Unknown |
c.508-1G>A; p.?; 3:158667075-158667075 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Unknown |
c.508-1G>A; p.?; 3:158667075-158667075 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Unknown |
c.508-1G>A; p.?; 3:158667075-158667075 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Unknown |
c.508-1G>A; p.?; 3:158667075-158667075 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Unknown |
c.280G>T; p.E94*; 3:158669523-158669523 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.405C>T; p.L135L; 3:158669098-158669098 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.405C>T; p.L135L; 3:158669098-158669098 |
urinary_tract; bladder | carcinoma | Substitution - coding silent |
c.250G>A; p.A84T; 3:158669553-158669553 |
breast | carcinoma | Substitution - Missense |
c.104A>T; p.Q35L; 3:158672375-158672375 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.241C>T; p.Q81*; 3:158669562-158669562 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.651G>T; p.K217N; 3:158666664-158666664 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.76G>T; p.G26W; 3:158672403-158672403 |
prostate | carcinoma; adenocarcinoma | Substitution - Missense |
c.328C>T; p.L110F; 3:158669475-158669475 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.401C>T; p.T134M; 3:158669102-158669102 |
thyroid | carcinoma; medullary_carcinoma | Substitution - Missense |
c.67T>C; p.Y23H; 3:158672412-158672412 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.196G>A; p.V66I; 3:158669607-158669607 |
skin | malignant_melanoma | Substitution - Missense |