| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 57111 |
Name | RAB25 |
Synonymous | RAB25, member RAS oncogene family;RAB25;RAB25, member RAS oncogene family |
Definition | ras-related protein Rab-25 |
Position | 1q22 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
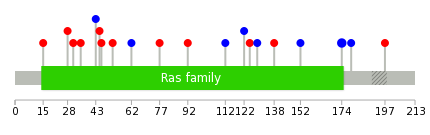 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
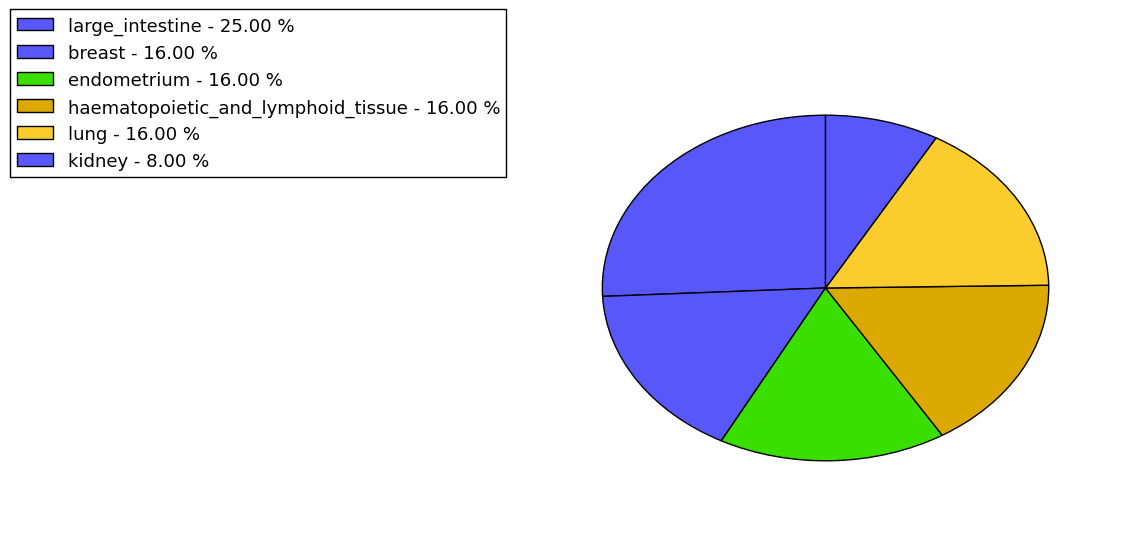 |
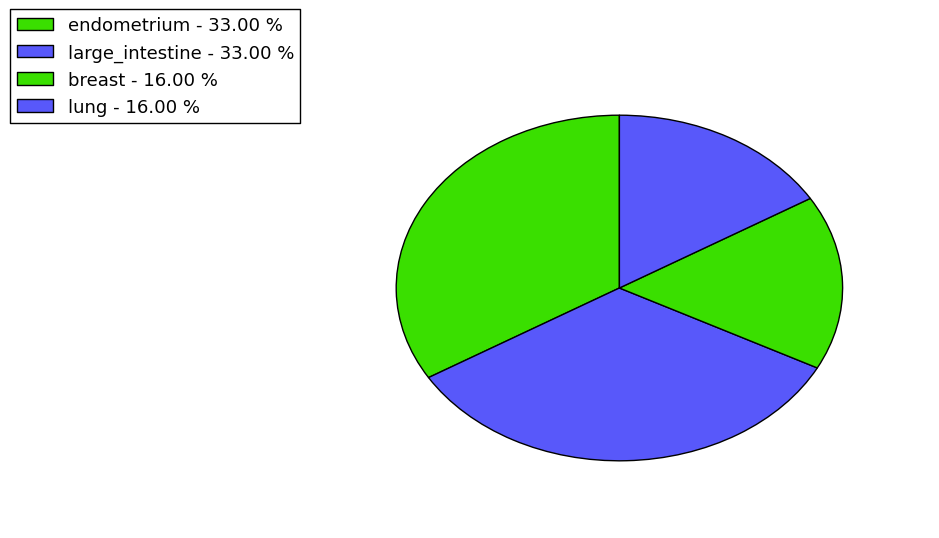 |
Loss of Function mutations compare to missense mutations | Top |
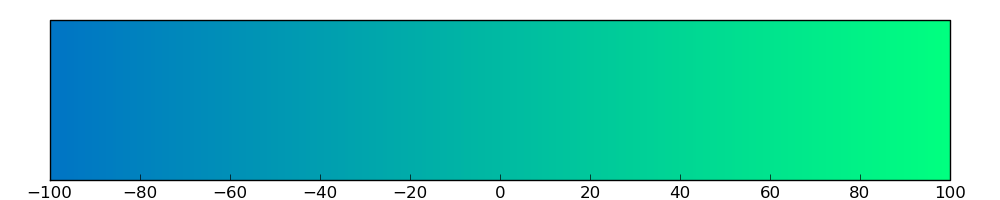 |
| The ratio of the loss-of-function mutations over missense mutations is 0.08. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.376C>T; p.L126F; 1:156068394-156068394 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.154G>C; p.E52Q; 1:156066009-156066009 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.375G>A; p.M125I; 1:156068393-156068393 |
breast | carcinoma | Substitution - Missense |
c.522G>T; p.L174L; 1:156069747-156069747 |
breast | carcinoma | Substitution - coding silent |
c.537G>A; p.A179A; 1:156070170-156070170 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.274G>A; p.A92T; 1:156068292-156068292 |
breast | carcinoma | Substitution - Missense |
c.487_497del11; p.S163fs*1; 1:156069712-156069722 |
large_intestine; colon | carcinoma; adenocarcinoma | Deletion - Frameshift |
c.186C>T; p.T62T; 1:156066041-156066041 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.522G>A; p.L174L; 1:156069747-156069747 |
lung; right_lower_lobe | carcinoma; adenocarcinoma | Substitution - coding silent |
c.229C>T; p.R77W; 1:156066084-156066084 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; acute_lymphoblastic_leukaemia | Substitution - Missense |
c.427G>T; p.E143*; 1:156068445-156068445 |
lung | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.135C>A; p.S45R; 1:156065990-156065990 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - Missense |
c.280C>T; p.L94L; 1:156068298-156068298 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.136C>T; p.R46C; 1:156065991-156065991 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.379G>A; p.V127M; 1:156068397-156068397 |
stomach | carcinoma; diffuse_adenocarcinoma | Substitution - Missense |
c.104G>A; p.R35Q; 1:156065959-156065959 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.104G>A; p.R35Q; 1:156065959-156065959 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.366C>T; p.I122I; 1:156068384-156068384 |
skin | malignant_melanoma | Substitution - coding silent |
c.43T>G; p.F15V; 1:156061431-156061431 |
large_intestine; colon | carcinoma | Substitution - Missense |
c.43T>G; p.F15V; 1:156061431-156061431 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.433C>T; p.R145*; 1:156068451-156068451 |
skin | malignant_melanoma | Substitution - Nonsense |
c.129C>T; p.H43H; 1:156065984-156065984 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.359C>T; p.A120V; 1:156068377-156068377 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.82G>A; p.G28R; 1:156065937-156065937 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.590G>A; p.G197D; 1:156070223-156070223 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.387C>T; p.N129N; 1:156068405-156068405 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.91A>G; p.N31D; 1:156065946-156065946 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.336G>A; p.K112K; 1:156068354-156068354 |
thyroid | carcinoma | Substitution - coding silent |
c.56-3C>T; p.?; 1:156065908-156065908 |
urinary_tract; bladder | carcinoma | Unknown |
c.412G>C; p.E138Q; 1:156068430-156068430 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.252G>A; p.A84A; 1:156068270-156068270 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.235C>T; p.R79*; 1:156066090-156066090 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.456A>T; p.G152G; 1:156069681-156069681 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |