| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 7447 |
Name | VSNL1 |
Synonymous | visinin-like 1;VSNL1;visinin-like 1 |
Definition | VLP-1|hippocalcin-like protein 3|visinin-like protein 1 |
Position | 2p24.3 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
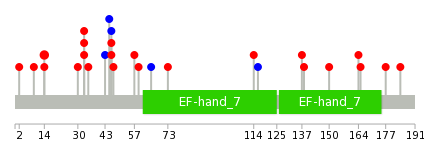 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
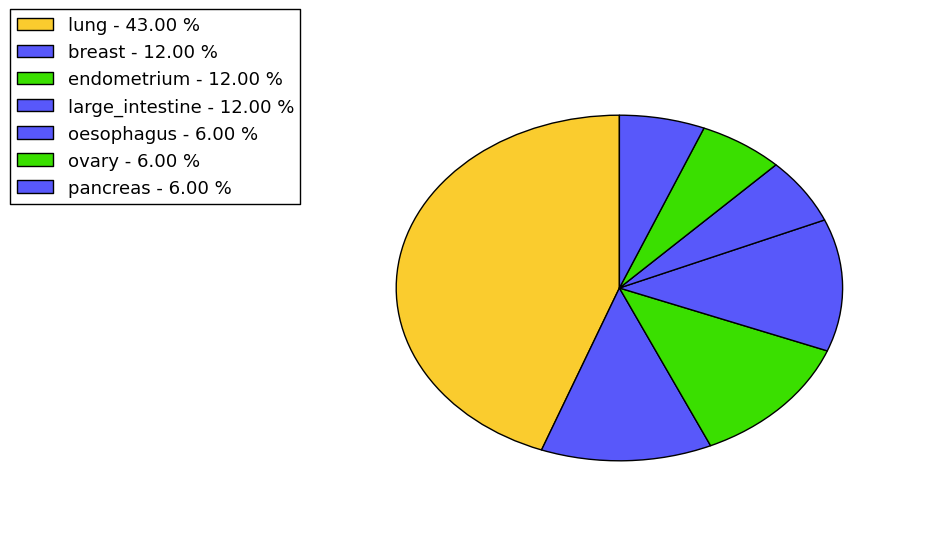 |
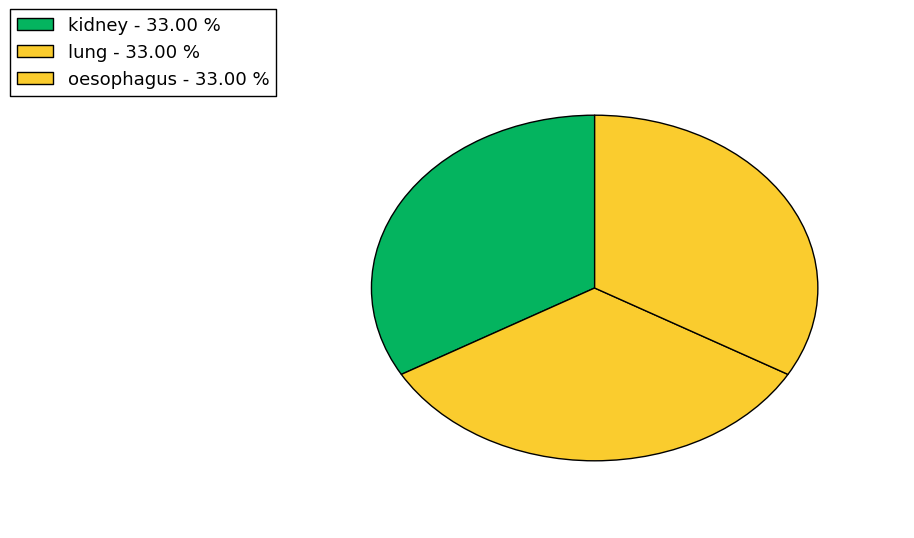 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.18. |
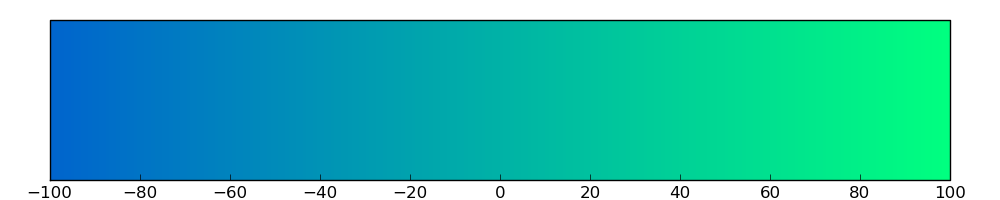
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.61G>T; p.E21*; 2:17592135-17592135 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Nonsense |
c.129A>G; p.L43L; 2:17592203-17592203 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - coding silent |
c.340G>A; p.G114S; 2:17649587-17649587 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.5G>T; p.G2V; 2:17592079-17592079 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.134T>C; p.L45P; 2:17592208-17592208 |
skin | malignant_melanoma | Substitution - Missense |
c.531C>A; p.S177R; 2:17655349-17655349 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.195C>A; p.A65A; 2:17649442-17649442 |
lung | carcinoma; adenocarcinoma | Substitution - coding silent |
c.271A>C; p.I91L; 2:17649518-17649518 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.136G>A; p.E46K; 2:17592210-17592210 |
skin | malignant_melanoma | Substitution - Missense |
c.413T>G; p.M138R; 2:17655231-17655231 |
breast | carcinoma | Substitution - Missense |
c.411G>T; p.M137I; 2:17655229-17655229 |
lung | carcinoma; small_cell_carcinoma | Substitution - Missense |
c.145C>T; p.Q49*; 2:17592219-17592219 |
skin | malignant_melanoma | Substitution - Nonsense |
c.97_98GG>AA; p.G33K; 2:17592171-17592172 |
skin | malignant_melanoma | Substitution - Missense |
c.25G>T; p.A9S; 2:17592099-17592099 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.140A>T; p.E47V; 2:17592214-17592214 |
skin | malignant_melanoma | Substitution - Missense |
c.42G>T; p.E14D; 2:17592116-17592116 |
ovary | carcinoma; serous_carcinoma | Substitution - Missense |
c.438G>A; p.T146T; 2:17655256-17655256 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.98G>A; p.G33E; 2:17592172-17592172 |
skin; trunk | malignant_melanoma | Substitution - Missense |
c.24delG; p.A9fs*4; 2:17592098-17592098 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; chronic_lymphocytic_leukaemia-small_lymphocytic_lymphoma | Deletion - Frameshift |
c.41A>G; p.E14G; 2:17592115-17592115 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.41A>G; p.E14G; 2:17592115-17592115 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.97G>A; p.G33R; 2:17592171-17592171 |
skin; mucosal | malignant_melanoma | Substitution - Missense |
c.218A>G; p.D73G; 2:17649465-17649465 |
oesophagus; lower_third | carcinoma; adenocarcinoma | Substitution - Missense |
c.550C>A; p.L184I; 2:17655368-17655368 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.169C>T; p.P57S; 2:17649416-17649416 |
skin; mucosal | malignant_melanoma | Substitution - Missense |
c.135C>T; p.L45L; 2:17592209-17592209 |
skin | malignant_melanoma | Substitution - coding silent |
c.103C>A; p.L35I; 2:17592177-17592177 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.263C>T; p.A88V; 2:17649510-17649510 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.136G>C; p.E46Q; 2:17592210-17592210 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.208C>T; p.R70*; 2:17649455-17649455 |
pancreas | carcinoma | Substitution - Nonsense |
c.176G>A; p.G59E; 2:17649423-17649423 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.490G>A; p.D164N; 2:17655308-17655308 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.448C>G; p.R150G; 2:17655266-17655266 |
lung; right_upper_lobe | carcinoma; adenocarcinoma | Substitution - Missense |
c.348C>T; p.I116I; 2:17649595-17649595 |
oesophagus; lower_third | carcinoma; adenocarcinoma | Substitution - coding silent |
c.89G>T; p.W30L; 2:17592163-17592163 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.493C>G; p.Q165E; 2:17655311-17655311 |
breast | carcinoma | Substitution - Missense |
c.175G>A; p.G59R; 2:17649422-17649422 |
skin | malignant_melanoma | Substitution - Missense |
c.357G>T; p.V119V; 2:17649604-17649604 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.138G>A; p.E46E; 2:17592212-17592212 |
skin | malignant_melanoma | Substitution - coding silent |