| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 85403 |
Name | EAF1 |
Synonymous | ELL associated factor 1;EAF1;ELL associated factor 1 |
Definition | ELL (eleven nineteen lysine-rich leukemia gene)-associated factor 1|ELL-associated factor 1 |
Position | 3p25.1 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
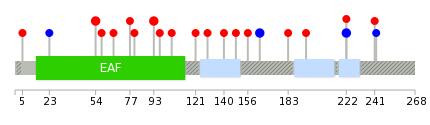 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
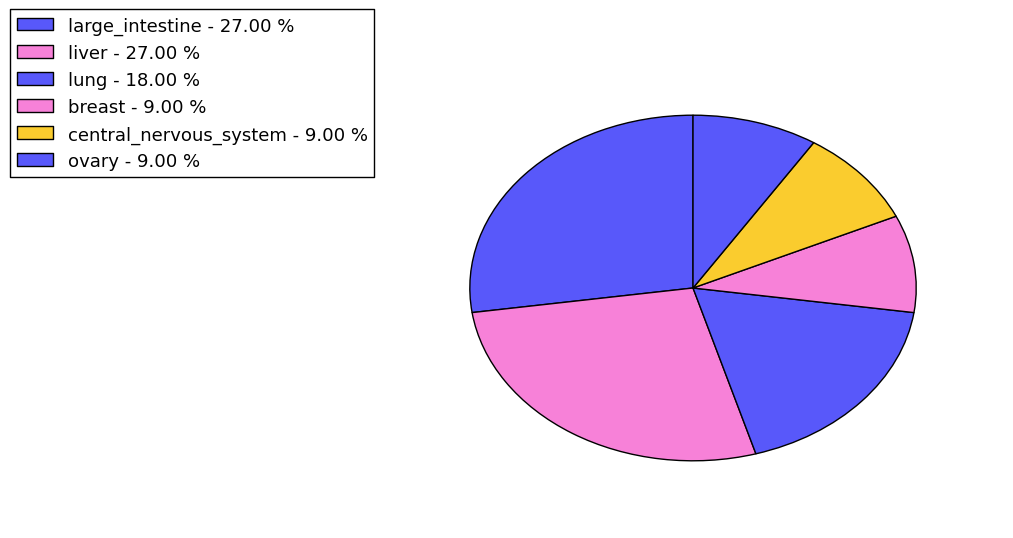 |
 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.08. |
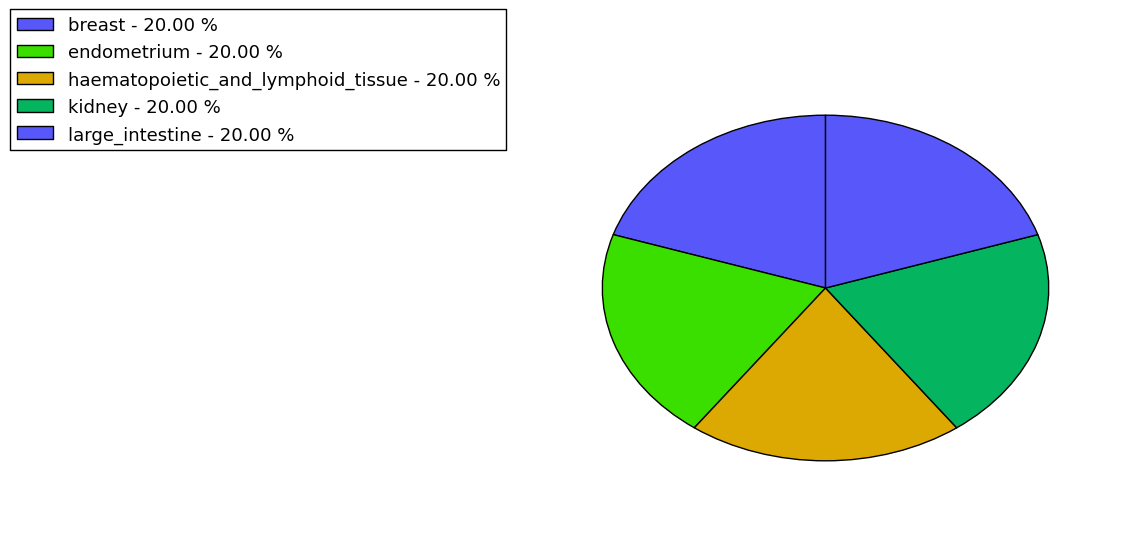
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.419C>T; p.P140L; 3:15434431-15434431 |
skin | malignant_melanoma | Substitution - Missense |
c.196C>G; p.P66A; 3:15430005-15430005 |
liver | carcinoma | Substitution - Missense |
c.467A>C; p.K156T; 3:15434479-15434479 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.728C>T; p.P243L; 3:15436543-15436543 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.238C>T; p.R80W; 3:15432126-15432126 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.69G>A; p.E23E; 3:15427848-15427848 |
breast | carcinoma | Substitution - coding silent |
c.442C>T; p.P148S; 3:15434454-15434454 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.666G>A; p.P222P; 3:15436481-15436481 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.666G>A; p.P222P; 3:15436481-15436481 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.722G>T; p.S241I; 3:15436537-15436537 |
central_nervous_system; brain | glioma | Substitution - Missense |
c.492C>T; p.P164P; 3:15434504-15434504 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - coding silent |
c.198T>G; p.P66P; 3:15430007-15430007 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.584C>G; p.S195C; 3:15436399-15436399 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.173T>C; p.V58A; 3:15429982-15429982 |
breast | carcinoma | Substitution - Missense |
c.492C>G; p.P164P; 3:15434504-15434504 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - coding silent |
c.492C>G; p.P164P; 3:15434504-15434504 |
kidney | other; neoplasm | Substitution - coding silent |
c.335+1G>A; p.?; 3:15432224-15432224 |
endometrium | carcinoma; endometrioid_carcinoma | Unknown |
c.361G>A; p.A121T; 3:15434373-15434373 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.290A>G; p.Y97C; 3:15432178-15432178 |
lung; right_lower_lobe | carcinoma; adenocarcinoma | Substitution - Missense |
c.314G>A; p.S105N; 3:15432202-15432202 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.386G>A; p.R129H; 3:15434398-15434398 |
prostate | carcinoma | Substitution - Missense |
c.160A>G; p.K54E; 3:15429969-15429969 |
liver | carcinoma | Substitution - Missense |
c.160A>G; p.K54E; 3:15429969-15429969 |
liver | carcinoma | Substitution - Missense |
c.548T>G; p.I183S; 3:15436363-15436363 |
ovary | carcinoma; serous_carcinoma | Substitution - Missense |
c.664C>T; p.P222S; 3:15436479-15436479 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.726G>A; p.R242R; 3:15436541-15436541 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.229G>A; p.G77R; 3:15432117-15432117 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.168T>C; p.D56D; 3:15429977-15429977 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.561_562insC; p.S188fs*12; 3:15436376-15436377 |
lung | carcinoma; adenocarcinoma | Insertion - Frameshift |
c.278A>G; p.D93G; 3:15432166-15432166 |
upper_aerodigestive_tract; mouth | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.13G>A; p.A5T; 3:15427792-15427792 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.278A>G; p.D93G; 3:15432166-15432166 |
upper_aerodigestive_tract; mouth | carcinoma | Substitution - Missense |