| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 9077 |
Name | DIRAS3 |
Synonymous | DIRAS family, GTP-binding RAS-like 3;DIRAS3;DIRAS family, GTP-binding RAS-like 3 |
Definition | GTP-binding protein Di-Ras3|distinct subgroup of the Ras family member 3|ras homolog gene family, member I|rho-related GTP-binding protein RhoI |
Position | 1p31 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
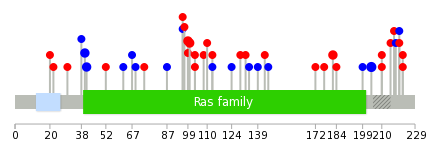 |
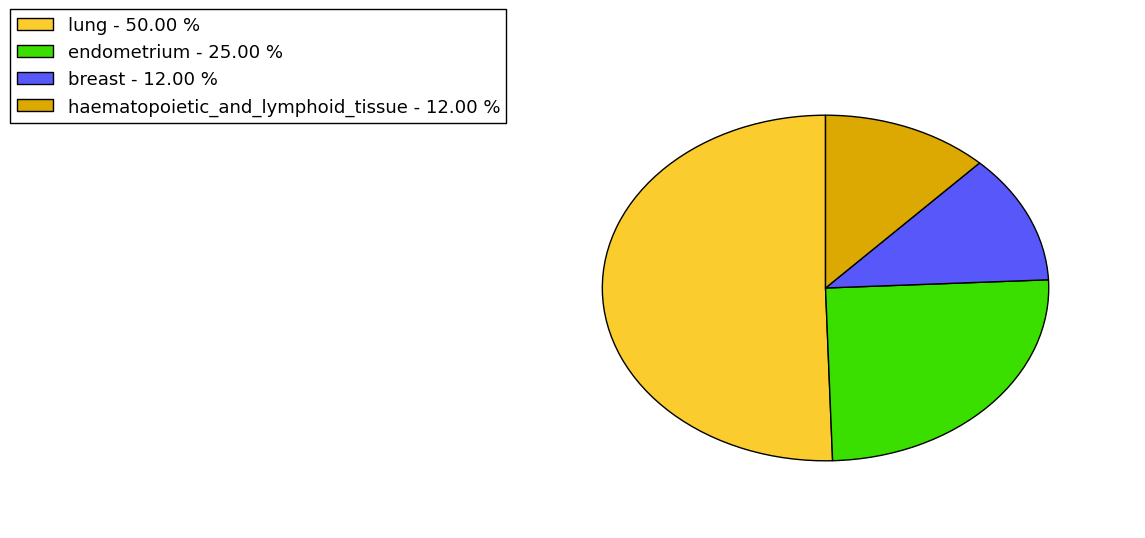
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
 |
 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.05. |
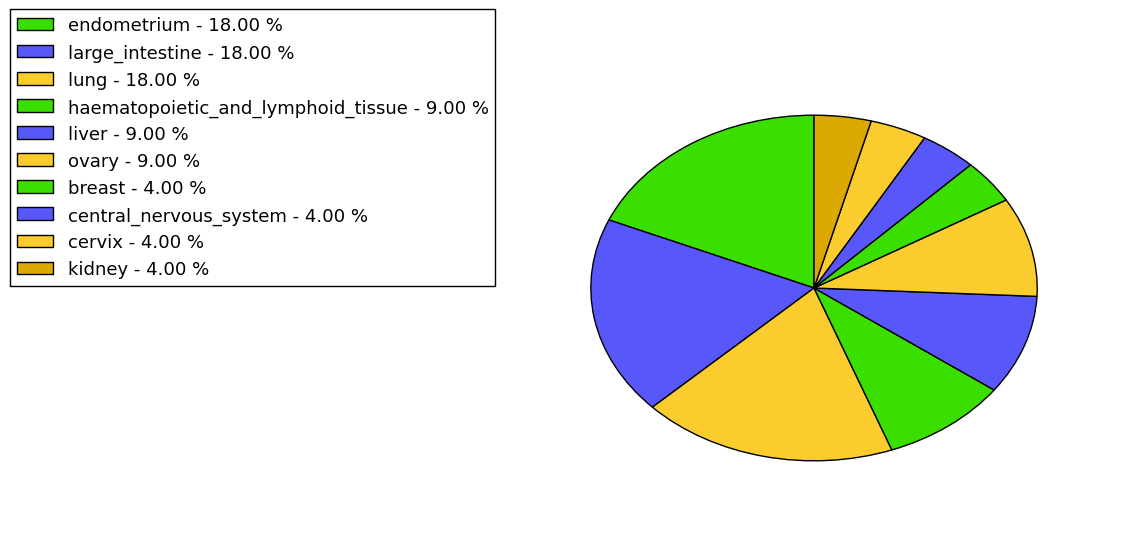
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.551C>G; p.T184S; 1:68046747-68046747 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.285C>T; p.G95G; 1:68047013-68047013 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.429T>G; p.H143Q; 1:68046869-68046869 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; follicular_lymphoma | Substitution - Missense |
c.155C>T; p.T52M; 1:68047143-68047143 |
upper_aerodigestive_tract; mouth | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.665T>A; p.L222Q; 1:68046633-68046633 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.417T>C; p.G139G; 1:68046881-68046881 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.199C>T; p.L67L; 1:68047099-68047099 |
skin | malignant_melanoma | Substitution - coding silent |
c.46C>G; p.R16G; 1:68047252-68047252 |
pleura | pulmonary_blastoma | Substitution - Missense |
c.261G>A; p.L87L; 1:68047037-68047037 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; Burkitt_lymphoma | Substitution - coding silent |
c.148A>G; p.K50E; 1:68047150-68047150 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.612C>T; p.T204T; 1:68046686-68046686 |
urinary_tract; bladder | carcinoma | Substitution - coding silent |
c.123G>T; p.V41V; 1:68047175-68047175 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.612C>T; p.T204T; 1:68046686-68046686 |
urinary_tract; bladder | carcinoma; transitional_cell_carcinoma | Substitution - coding silent |
c.612C>T; p.T204T; 1:68046686-68046686 |
urinary_tract; bladder | carcinoma | Substitution - coding silent |
c.114C>T; p.Y38Y; 1:68047184-68047184 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.123G>A; p.V41V; 1:68047175-68047175 |
skin | malignant_melanoma | Substitution - coding silent |
c.288C>T; p.D96D; 1:68047010-68047010 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.649A>G; p.N217D; 1:68046649-68046649 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.597G>A; p.K199K; 1:68046701-68046701 |
skin | malignant_melanoma | Substitution - coding silent |
c.337G>A; p.V113I; 1:68046961-68046961 |
salivary_gland | carcinoma; adenoid_cystic_carcinoma | Substitution - Missense |
c.221A>T; p.Y74F; 1:68047077-68047077 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; follicular_lymphoma | Substitution - Missense |
c.177C>T; p.S59S; 1:68047121-68047121 |
oesophagus | carcinoma; adenocarcinoma | Substitution - coding silent |
c.522T>A; p.N174K; 1:68046776-68046776 |
skin | malignant_melanoma | Substitution - Missense |
c.308G>T; p.R103L; 1:68046990-68046990 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.296G>A; p.R99H; 1:68047002-68047002 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.59T>A; p.L20Q; 1:68047239-68047239 |
ovary | carcinoma; serous_carcinoma | Substitution - Missense |
c.296G>A; p.R99H; 1:68047002-68047002 |
skin; mucosal | malignant_melanoma | Substitution - Missense |
c.296G>C; p.R99P; 1:68047002-68047002 |
central_nervous_system; brain | glioma; astrocytoma_Grade_IV | Substitution - Missense |
c.402C>A; p.I134I; 1:68046896-68046896 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.322C>T; p.R108W; 1:68046976-68046976 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.514G>A; p.E172K; 1:68046784-68046784 |
skin | malignant_melanoma | Substitution - Missense |
c.286G>A; p.D96N; 1:68047012-68047012 |
skin; acral | malignant_melanoma | Substitution - Missense |
c.298G>A; p.A100T; 1:68047000-68047000 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.298G>A; p.A100T; 1:68047000-68047000 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.298G>A; p.A100T; 1:68047000-68047000 |
NS | NS | Substitution - Missense |
c.298G>A; p.A100T; 1:68047000-68047000 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.298G>A; p.A100T; 1:68047000-68047000 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.298G>A; p.A100T; 1:68047000-68047000 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.435C>T; p.F145F; 1:68046863-68046863 |
skin | malignant_melanoma | Substitution - coding silent |
c.386C>T; p.A129V; 1:68046912-68046912 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.660G>A; p.E220E; 1:68046638-68046638 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.186C>T; p.F62F; 1:68047112-68047112 |
lung; right_upper_lobe | carcinoma; adenocarcinoma | Substitution - coding silent |
c.610A>G; p.T204A; 1:68046688-68046688 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.654C>T; p.T218T; 1:68046644-68046644 |
lung; right_upper_lobe | carcinoma; adenocarcinoma | Substitution - coding silent |
c.64G>A; p.A22T; 1:68047234-68047234 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.690A>G; p.*230W; 1:68046608-68046608 |
stomach | carcinoma; adenocarcinoma | Nonstop extension |
c.240C>A; p.C80*; 1:68047058-68047058 |
stomach | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.628G>T; p.E210*; 1:68046670-68046670 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.660G>T; p.E220D; 1:68046638-68046638 |
breast | carcinoma | Substitution - Missense |
c.207C>T; p.T69T; 1:68047091-68047091 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.330C>A; p.H110Q; 1:68046968-68046968 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.69G>A; p.L23L; 1:68047229-68047229 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.643A>G; p.M215V; 1:68046655-68046655 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.120C>T; p.V40V; 1:68047178-68047178 |
urinary_tract; bladder | carcinoma | Substitution - coding silent |
c.120C>T; p.V40V; 1:68047178-68047178 |
urinary_tract; bladder | carcinoma; transitional_cell_carcinoma | Substitution - coding silent |
c.339C>T; p.V113V; 1:68046959-68046959 |
breast | carcinoma | Substitution - coding silent |
c.370C>T; p.L124L; 1:68046928-68046928 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.394G>A; p.E132K; 1:68046904-68046904 |
skin; upper_arm | malignant_melanoma | Substitution - Missense |
c.628G>C; p.E210Q; 1:68046670-68046670 |
cervix | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.90G>T; p.K30N; 1:68047208-68047208 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.289G>A; p.G97S; 1:68047009-68047009 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.63C>T; p.P21P; 1:68047235-68047235 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.628G>A; p.E210K; 1:68046670-68046670 |
haematopoietic_and_lymphoid_tissue; lymph_node | lymphoid_neoplasm; follicular_lymphoma | Substitution - Missense |
c.628G>A; p.E210K; 1:68046670-68046670 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.664C>G; p.L222V; 1:68046634-68046634 |
ovary | other; neoplasm | Substitution - Missense |
c.308G>A; p.R103H; 1:68046990-68046990 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.545C>T; p.A182V; 1:68046753-68046753 |
liver | carcinoma | Substitution - Missense |
c.545C>T; p.A182V; 1:68046753-68046753 |
liver | carcinoma | Substitution - Missense |
c.279G>A; p.K93K; 1:68047019-68047019 |
skin | malignant_melanoma | Substitution - coding silent |
c.529T>G; p.F177V; 1:68046769-68046769 |
prostate | carcinoma; adenocarcinoma | Substitution - Missense |
c.204G>A; p.P68P; 1:68047094-68047094 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.116G>A; p.R39H; 1:68047182-68047182 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |