| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 9368 |
Name | SLC9A3R1 |
Synonymous | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1;SLC9A3R1;solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
Definition | Na(+)/H(+) exchange regulatory cofactor NHE-RF1|Na+/H+ exchange regulatory co-factor|ezrin-radixin-moesin binding phosphoprotein-50|ezrin-radixin-moesin-binding phosphoprotein 50|regulatory cofactor of Na(+)/H(+) exchanger|solute carrier family 9 (sodium/ |
Position | 17q25.1 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
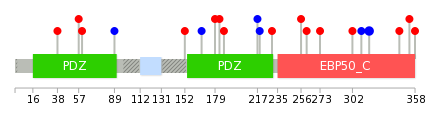 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
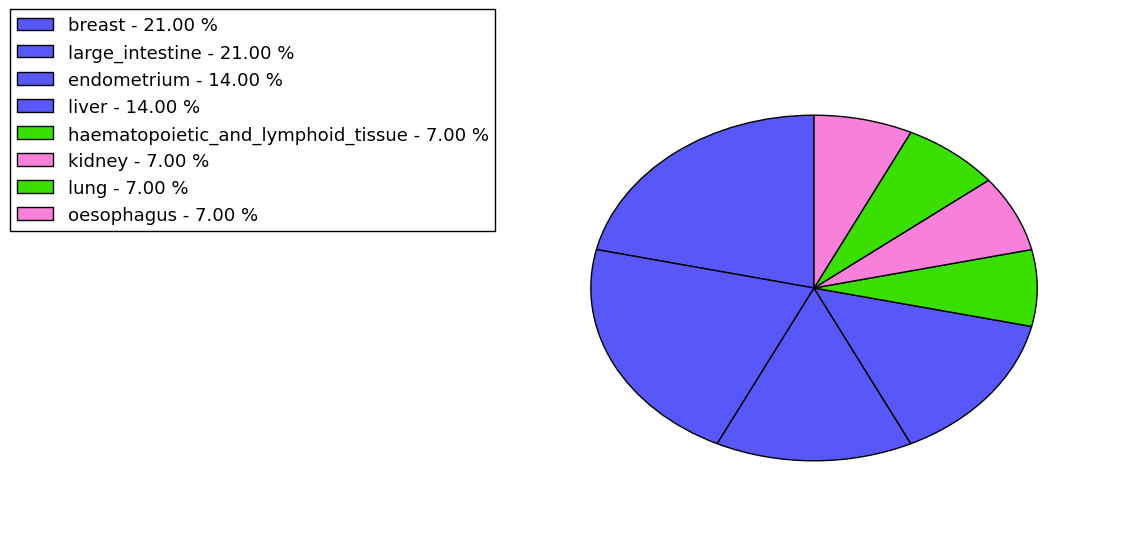 |
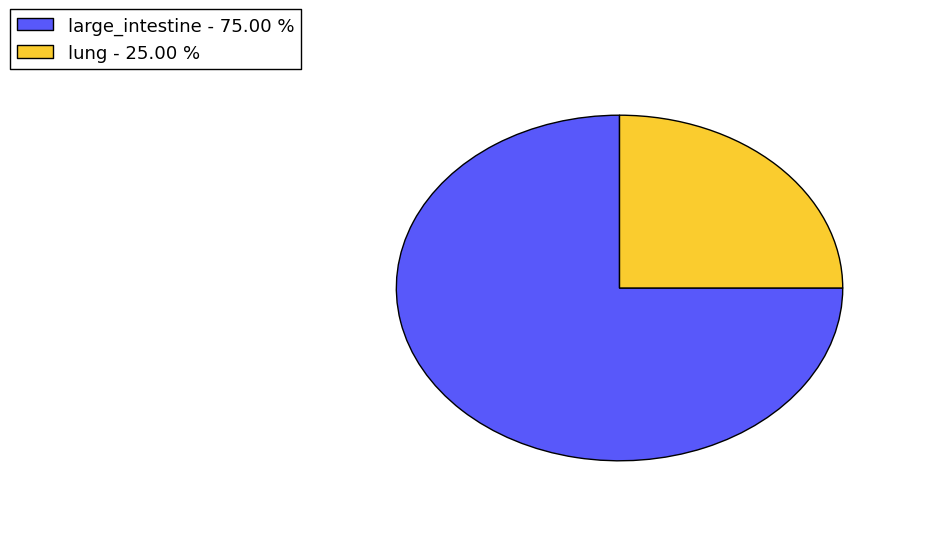 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.14. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.179T>G; p.V60G; 17:74749025-74749025 |
liver | carcinoma | Substitution - Missense |
c.930G>A; p.A310A; 17:74768509-74768509 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.415G>C; p.E139Q; 17:74749261-74749261 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.951C>T; p.S317S; 17:74768530-74768530 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.951C>T; p.S317S; 17:74768530-74768530 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.905G>C; p.S302T; 17:74768484-74768484 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - Missense |
c.799-1G>A; p.?; 17:74768146-74768146 |
pancreas | carcinoma | Unknown |
c.1073T>C; p.L358P; 17:74768652-74768652 |
breast | carcinoma | Substitution - Missense |
c.799-1G>A; p.?; 17:74768146-74768146 |
pancreas | carcinoma; ductal_carcinoma | Unknown |
c.301A>T; p.K101*; 17:74749147-74749147 |
kidney | carcinoma; papillary_renal_cell_carcinoma | Substitution - Nonsense |
c.560C>G; p.P187R; 17:74762130-74762130 |
breast | carcinoma | Substitution - Missense |
c.940A>G; p.T314A; 17:74768519-74768519 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.767C>A; p.P256H; 17:74766945-74766945 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.170A>G; p.D57G; 17:74749016-74749016 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.651C>A; p.S217S; 17:74763414-74763414 |
thyroid | carcinoma | Substitution - coding silent |
c.1057G>A; p.E353K; 17:74768636-74768636 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.454C>T; p.P152S; 17:74762024-74762024 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.501C>T; p.N167N; 17:74762071-74762071 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.688G>A; p.V230M; 17:74763451-74763451 |
skin | malignant_melanoma | Substitution - Missense |
c.112T>C; p.Y38H; 17:74748958-74748958 |
breast | carcinoma | Substitution - Missense |
c.782A>G; p.N261S; 17:74766960-74766960 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.787G>T; p.E263*; 17:74766965-74766965 |
oesophagus; lower_third | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.666C>T; p.G222G; 17:74763429-74763429 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.371C>T; p.A124V; 17:74749217-74749217 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.886G>A; p.E296K; 17:74768234-74768234 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.818T>C; p.L273P; 17:74768166-74768166 |
liver | carcinoma | Substitution - Missense |
c.657C>T; p.I219I; 17:74763420-74763420 |
lung; right_lower_lobe | carcinoma; adenocarcinoma | Substitution - coding silent |
c.267G>A; p.L89L; 17:74749113-74749113 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.1030C>T; p.P344S; 17:74768609-74768609 |
skin | malignant_melanoma | Substitution - Missense |
c.548A>T; p.D183V; 17:74762118-74762118 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.535A>G; p.I179V; 17:74762105-74762105 |
stomach | adenocarcinoma | Substitution - Missense |