| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 3688 |
Name | ITGB1 |
Synonymous | CD29|FNRB|GPIIA|MDF2|MSK12|VLA-BETA|VLAB;integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12);ITGB1;integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
Definition | glycoprotein IIa|integrin VLA-4 beta subunit|integrin beta-1|very late activation protein, beta polypeptide |
Position | 10p11.2 |
Gene Type | protein-coding |
Gene Regulation: | Upstream microRNAs Transcription Factors Post Transcriptional Modification From dbPTM Methylation Profile From DiseaseMeth database |
Experimentally verified miRNA from miRTarBase [Top] | |||
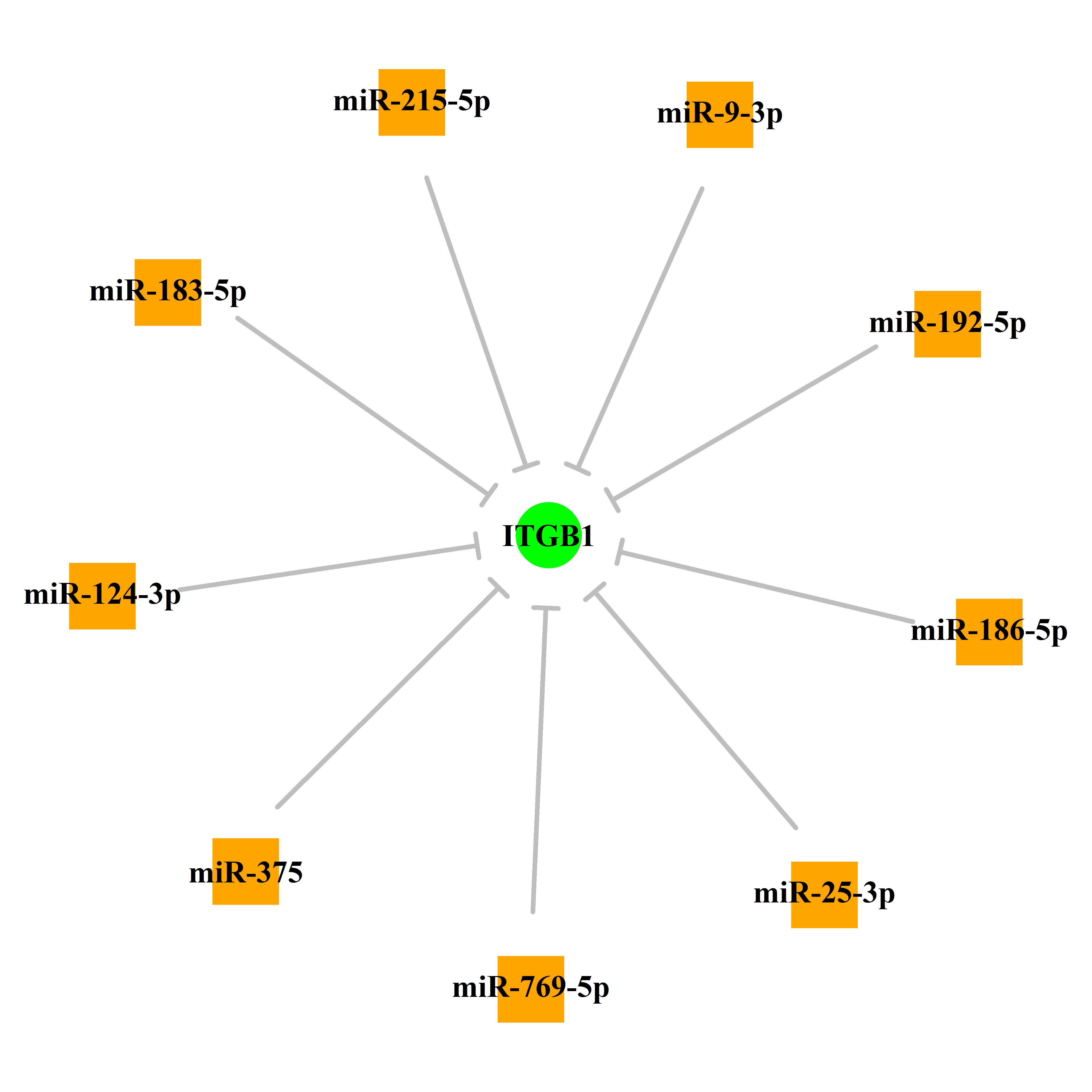 |
Transcription Factors [Top] |
|
Regulation From TransFac database |
alpha8-integrin(h) + LAP1(mo) + beta1-integrin(h) <==> LAP1(mo):alpha8-integrin(h):beta1-integrin(h) (binding) | LAP1(mo) + CD51(h) + beta1-integrin(h) <==> LAP1(mo):CD51(h):beta1-integrin(h) (binding) | TGFbeta1(h) + LAP1(h) + beta1-integrin(h) <==> TGFbeta1(h):LAP1(h):beta1-integrin(h) (binding) | LAP1(h) + CD51(h) + beta1-integrin(h) <==> LAP1(h):CD51(h):beta1-integrin(h) (binding) | beta1-integrin(h) + RACK1(h) <==> beta1-integrin(h):RACK1(h) (binding) | beta1-integrin(h) + ICAP-1alpha(h) <==> beta1-integrin(h):ICAP-1alpha(h) (binding) | beta1-integrin(h) + alpha-actinin(c) <==> beta1-integrin(h):alpha-actinin(c) (binding) | beta1-integrin(h) --> Pyk2-isoform1(h) (activation; phosphorylation) | beta1-integrin(h) + IGF-1R(h) <==> beta1-integrin(h):IGF-1R(h) (binding) | beta1-integrin(h) + IGF-1R(ha) <==> beta1-integrin(h):IGF-1R(ha) (binding) | beta1-integrin(h) + IRS-1(ha) <==> beta1-integrin(h):IRS-1(ha) (binding) | beta1-integrin(h) + Gab-1(ha) <==> beta1-integrin(h):Gab-1(ha) (binding) | beta1-integrin(h) --> Gab-1(ha){pY} (increase of phosphorylation) | beta1-integrin(h) --> Gab-1(ha):SHP-2(ha){pY542} (increase of phosphorylation) | beta1-integrin(h) + MK(h) <==> beta1-integrin(h):MK(h) (binding) | Lrp6(m) + beta1-integrin(h) <==> Lrp6(m):beta1-integrin(h) (binding) | RPTPzeta(r) + beta1-integrin(h) <==> RPTPzeta(r):beta1-integrin(h) (binding) | caveolin-1(h) + beta1-integrin(h) <==> caveolin-1(h):beta1-integrin(h) (binding) | beta1-integrin(h) + LAMA5(h) <==> beta1-integrin(h):LAMA5(h) (binding) | IGF-1R-beta-p97(h){pY} + beta1-integrin(h) <==> beta1-integrin(h):IGF-1R-beta-p97(h){pY} (binding) | beta1-integrin(h) + paxillin(h){pY} <==> beta1-integrin(h):paxillin(h){pY} (binding) | beta1-integrin(h) + FAK1(h){pY} <==> beta1-integrin(h):FAK1(h){pY} (binding) | beta1-integrin(h) + alpha-actinin(h) <==> beta1-integrin(h):alpha-actinin(h) (binding) | PKCalpha(h) + beta1-integrin(h) <==> PKCalpha(h):beta1-integrin(h) (binding) | beta1-integrin(h) --> FAK(h){pY} (increase of phosphorylation) | beta1-integrin(h) --> HEF1(h){pY} (increase of phosphorylation) | beta1-integrin(h) --> c-Cbl(h){pY} (increase of phosphorylation) | beta1-integrin(h) --> c-Cbl(h):CrkL(m.s.) (increase of binding) | beta1-integrin(h) --> HEF1(h):CrkL(m.s.) (increase of binding) | beta1-integrin(h) --> Cas(h){pY} (increase of phosphorylation) | beta1-integrin(h) --> Cas(h):Fyn(h){pY} (increase of binding) | beta1-integrin(h) --> SHP-2(h):Cas(h) (increase of binding) | beta1-integrin(h) --> HEF1(h):CrkL(h) (increase of binding) | beta1-integrin(h) --> Cas(h):CrkL(h) (increase of binding) | beta1-integrin(h) --> Lyn(h){pY} (increase of phosphorylation) | beta1-integrin(h) + TGC(h) <==> beta1-integrin(h):TGC(h) (binding) | Fibronectin-p42(m.s.) + TGC(h) + beta1-integrin(h) <==> Fibronectin-p42(m.s.):beta1-integrin(h):TGC(h) (binding) | beta1-integrin(h) + fibronectin(m.s.) <==> beta1-integrin(h):fibronectin(m.s.) (binding) | beta1-integrin(h) + Fibronectin-p42(m.s.) <==> beta1-integrin(h):Fibronectin-p42(m.s.) (binding) | uPAR(h) + beta1-integrin(h) <==> uPAR(h):beta1-integrin(h) (binding) | ErbB2(m.s.) + beta1-integrin(h) <==> ErbB2(m.s.):beta1-integrin(h) (binding) | FAK1(h) + beta1-integrin(h) <==> FAK1(h):beta1-integrin(h) (binding) | alpha4-integrin(h) + beta1-integrin(h) <==> alpha4-integrin(h):beta1-integrin(h) (binding) | Itgb1(h) --> beta1-integrin(h) (expression). | LAT(h) --> beta1-integrin(h) (activation) | IGF-1(h) --> IRS-1(ha){pY} (increase of phosphorylation) | beta1A-integrin(h) + MIBP(h) <==> beta1A-integrin(h):MIBP(h) (binding) | beta1A-integrin(h) + ICAP-1(r) <==> beta1A-integrin(h):ICAP-1(r) (binding) | beta1A-integrin(h) + melusin(r) <==> beta1A-integrin(h):melusin(r) (binding) | beta1A-integrin(h) + talin(h) <==> beta1A-integrin(h):talin(h) (binding) | beta1A-integrin(h) + Syk(h) <==> beta1A-integrin(h):Syk(h) (binding) | beta1A-integrin(h) --> EGFR(m){pY} (increase of phosphorylation) | beta1A-integrin(h) --> Src(m){pY424} (increase of phosphorylation) | beta1A-integrin(h) --> Cas(m){pY} (increase of phosphorylation) | beta1A-integrin(h) + TGC(ha) <==> beta1A-integrin(h):TGC(ha) (binding) | Itgb1(h) --> beta1A-integrin(h) (expression). | beta1D-integrin(h) + MIBP(h) <==> beta1D-integrin(h):MIBP(h) (binding) | beta1D-integrin(h) + melusin(r) <==> beta1D-integrin(h):melusin(r) (binding) | beta1D-integrin(h) + talin(h) <==> beta1D-integrin(h):talin(h) (binding) | beta1D-integrin(h) + TGC(ha) <==> beta1D-integrin(h):TGC(ha) (binding) | Itgb1(h) --> beta1D-integrin(h) (expression). | Itgb1(h) --> beta1B-integrin(h) (expression). | Itgb1(h) --> beta1C-integrin(h) (expression). | Itgb1(h) --> beta1C-2-integrin(h) (expression). |
|---|
Post Transcriptional Modification [Top] | |||
|
Location (AA) |
PTM type |
Literature |
Database |
50 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 94 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 97 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 212 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 269 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 363 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 406 | N-linked (GlcNAc...). | Swiss-Prot 53.0 | 417 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 481 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 520 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 584 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 669 | N-linked (GlcNAc...) (Potential). | Swiss-Prot 53.0 | 783 | Phosphotyrosine (By similarity). | Swiss-Prot 53.0 | 788 | Phosphothreonine | 14660602 | Phospho.ELM 6.0 | 789 | Phosphothreonine | 14660602 | Phospho.ELM 6.0 |
|---|---|---|---|
Methylation Profile [Top] | ||
|
Chromosome |
Location |
Source |
|---|---|---|
chr10 |
Promoter: 33286704 - 33288704 | Agilent_014791 |
chr10 |
Promoter: 33286328 - 33288328 | Agilent_014791 |
chr10 |
Promoter: 33286704 - 33288704 | GoldenGate_Methylation_Cancer_Panel_I |
chr10 |
Promoter: 33286328 - 33288328 | GoldenGate_Methylation_Cancer_Panel_I |
chr10 |
Promoter: 33286328 - 33288328 | GSE27584 |
chr10 |
Promoter: 33286704 - 33288704 | GSE27584 |
chr10 |
Promoter: 33286704 - 33288704 | Agilent_014791_44K |
chr10 |
Promoter: 33286328 - 33288328 | Agilent_014791_44K |