| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 6294 |
Name | SAFB |
Synonymous | HAP|HET|SAF-B1|SAFB1;scaffold attachment factor B;SAFB;scaffold attachment factor B |
Definition | HSP27 ERE-TATA-binding protein|HSP27 estrogen response element-TATA box-binding protein|Hsp27 ERE-TATA binding protein|SAB-B1|SAF-B|glutathione S-transferase fusion protein|heat-shock protein (HSP27) estrogen response element and TATA box-binding protein| |
Position | 19p13.3-p13.2 |
Gene Type | protein-coding |
Gene Regulation: | Upstream microRNAs Transcription Factors Post Transcriptional Modification From dbPTM Methylation Profile From DiseaseMeth database |
Experimentally verified miRNA from miRTarBase [Top] | |||
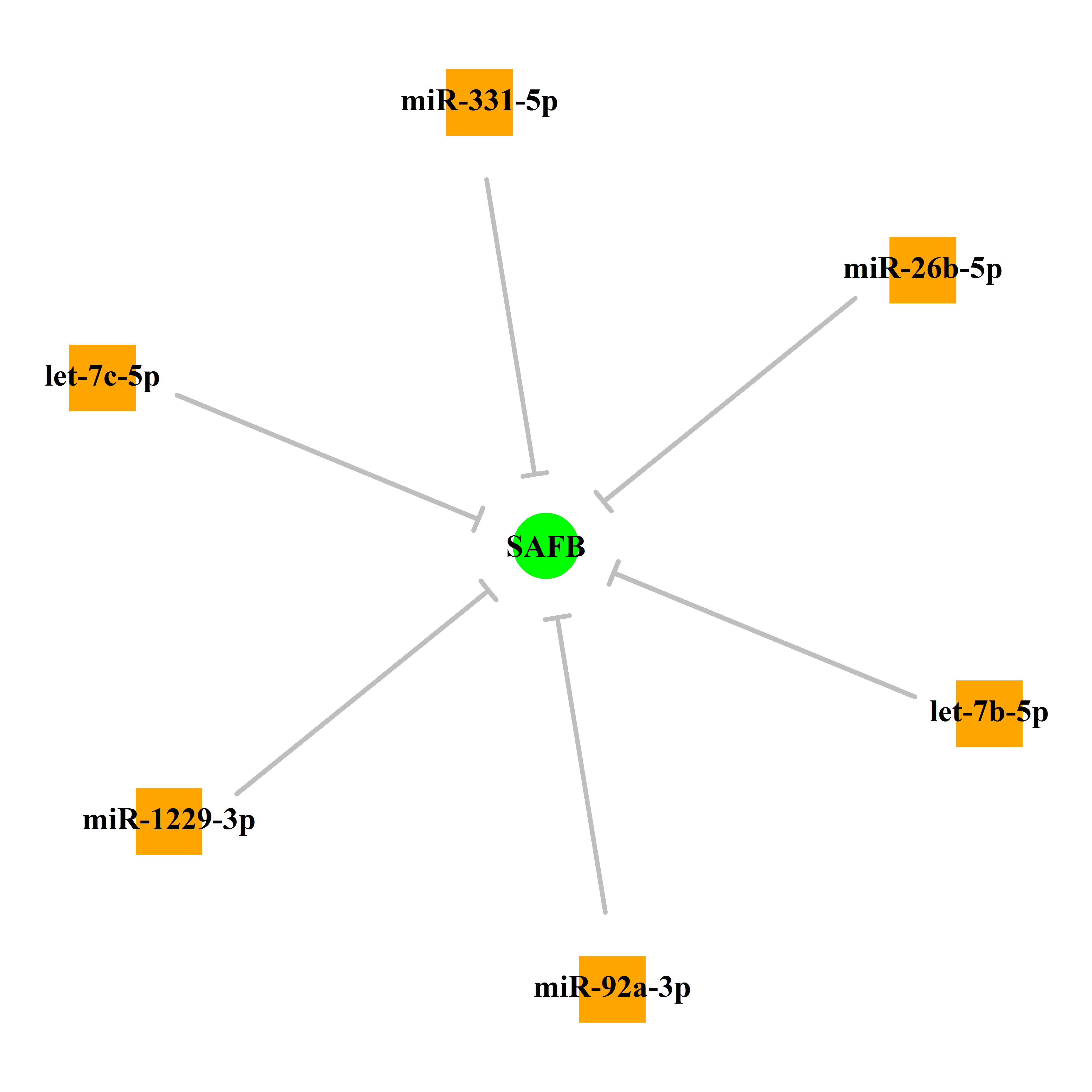 |
Transcription Factors [Top] |
|
Regulation From TransFac database |
Ku70(h) + Ccte(h) + Pds5A(h) + PPIL1(h) + eIF-3gamma(h) + SAFB1(h) + Wapl(h) + Rad21(h) + SMC-3(h) + SMC1(h) + CAPER-alpha(h) + CNOT3(h) + NCoA-5(h) + SIAHBP1(h) + FBP(h) + HYPA(h) + SKIP(h) + SRm300(h) + SRm160(h) + p54nrb/NonO(h) + rbm25(h) + rbm8A(h) + Pinin(h) + Prp19(h) + cdc5L(h) + Snrp116(h) + Prpf8(h) + Sf3b-4(h) + Sf3b-2(h) + hnRNPM(h) + hnRNP-G(h) <==> Ku70(h):Ccte(h):Pds5A(h):PPIL1(h):eIF-3gamma(h):SAFB1(h):Wapl(h):Rad21(h):SMC-3(h):SMC1(h):CAPER-alpha(h):CNOT3(h):NCoA-5(h):SIAHBP1(h):FBP(h):HYPA(h):SKIP(h):SRm300(h):SRm160(h):p54nrb/NonO(h):rbm25(h):rbm8A(h):Pinin(h):Prp19(h):cdc5L(h):Snrp116(h):Prpf8(h):Sf3b-4(h):Sf3b-2(h):hnRNPM(h):hnRNP-G(h) (binding) | SAFB1(h) + Mi2-alpha(h) <==> SAFB1(h):Mi2-alpha(h) (binding) | SAFB(h) --> SAFB1(h) (expression). | SAFB(h) --> SAFB1(h) (expression). |
|---|
Post Transcriptional Modification [Top] | |||
|
Location (AA) |
PTM type |
Literature |
Database |
288 | Phosphoserine. | Swiss-Prot 53.0 | 344 | Phosphoserine (By similarity). | Swiss-Prot 53.0 | 384 | Phosphoserine. | Swiss-Prot 53.0 | 589 | Phosphoserine. | Swiss-Prot 53.0 | 601 | Phosphoserine. | Swiss-Prot 53.0 | 604 | Phosphoserine. | Swiss-Prot 53.0 | 808 | Phosphoserine. | Swiss-Prot 53.0 | 604 | Phosphoserine | 15302935;17081983 | Phospho.ELM 6.0 | 288 | Phosphoserine | 15302935 | Phospho.ELM 6.0 | 601 | Phosphoserine | 15302935 | Phospho.ELM 6.0 | 416 | Phosphothreonine | 15302935 | Phospho.ELM 6.0 | 20 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 21 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 23 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 32 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 194 | Phosphothreonine | 17081983 | Phospho.ELM 6.0 | 234 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 344 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 384 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 555 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 589 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 621 | Phosphoserine | 17081983 | Phospho.ELM 6.0 | 808 | Phosphoserine | 17081983 | Phospho.ELM 6.0 |
|---|---|---|---|
Methylation Profile [Top] | ||
|
Chromosome |
Location |
Source |
|---|---|---|
chr19 |
Promoter: 5572664 - 5574664 | GSE27584 |
chr19 |
Promoter: 5572664 - 5574664 | Agilent_014791_44K |