| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 8454 |
Name | CUL1 |
Synonymous | -;cullin 1;CUL1;cullin 1 |
Definition | CUL-1|cullin-1 |
Position | 7q36.1 |
Gene Type | protein-coding |
Gene Regulation: | Upstream microRNAs Transcription Factors Post Transcriptional Modification From dbPTM Methylation Profile From DiseaseMeth database |
Experimentally verified miRNA from miRTarBase [Top] | |||
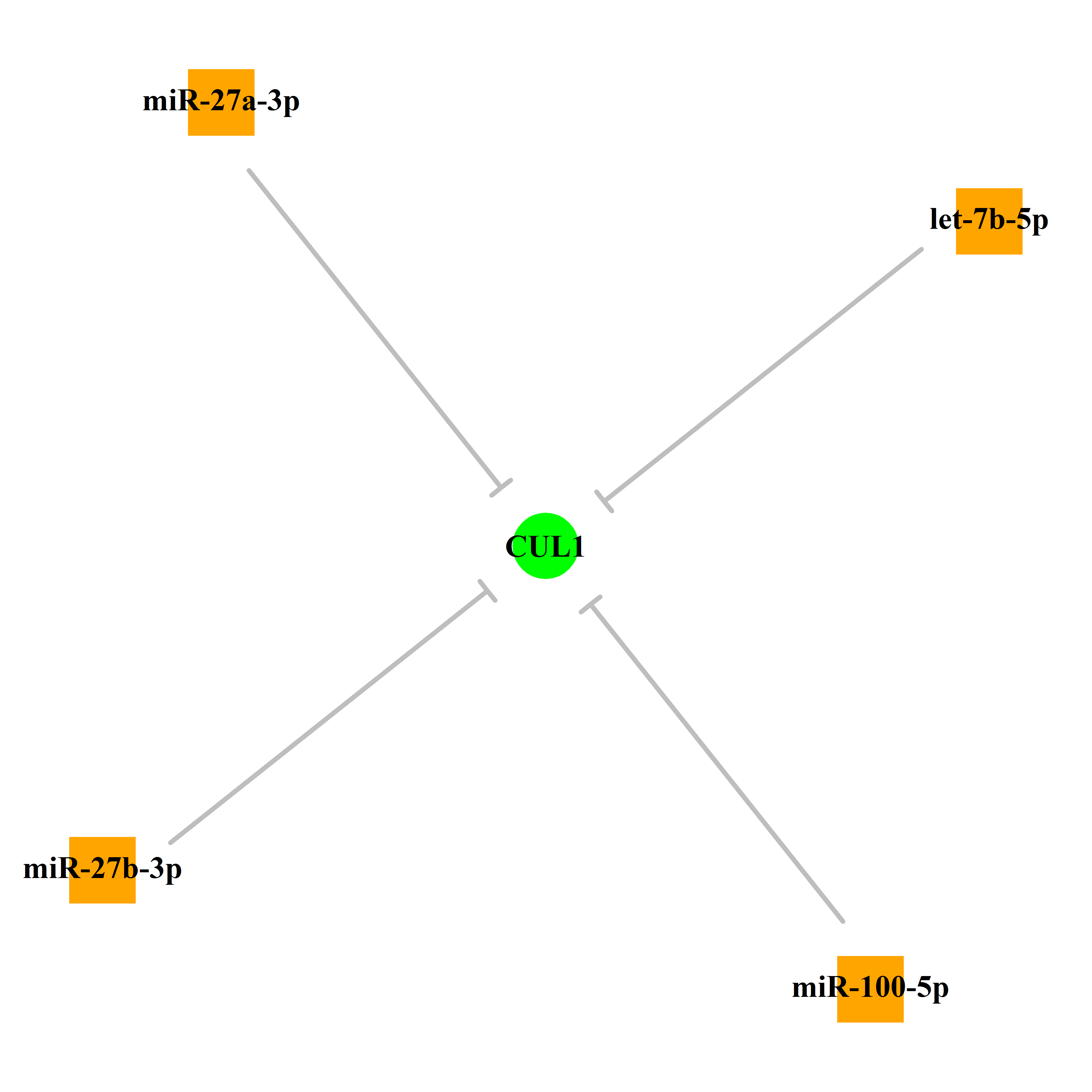 |
Transcription Factors [Top] |
|
Regulation From TransFac database |
Cul-1(h) + Skp1(h) <==> Cul-1(h):Skp1(h) (binding) | Cul-1(h) + Roc1(h) <==> Roc1(h):Cul-1(h) (binding) | beta-TrCP1(h) + Cul-1(h) + Skp1(h) <==> beta-TrCP1(h):Cul-1(h):Skp1(h) (binding) | Cul-1(h) + Fbx4(h) + Skp1(h) <==> Cul-1(h):Fbx4(h):Skp1(h) (binding) | Cul-1(h) + Fbx7(h) + Skp1(h) <==> Cul-1(h):Fbx7(h):Skp1(h) (binding) | Cul-1(h) + Fbw2(h) + Skp1(h) <==> Cul-1(h):Fbw2(h):Skp1(h) (binding) | Cul-1(h) + Fbl3a(h) + Skp1(h) <==> Cul-1(h):Fbl3a(h):Skp1(h) (binding) | Cul-1(h) + HOS(h) + Roc1(h) + Skp1(h) <==> Cul-1(h):HOS(h):Roc1(h):Skp1(h) (binding) | Cul-1(h) + Roc1(h) + Skp1(h) <==> Cul-1(h):Roc1(h):Skp1(h) (binding) | Cul-1(h) + HOS(h) + Skp1(h) <==> Cul-1(h):HOS(h):Skp1(h) (binding) | Cul-1(h) + beta-TrCP1(m) + Skp1(h) <==> Cul-1(h):beta-TrCP1(m):Skp1(h) (binding) | Cul-1(h) + beta-TrCP1(m) + IkappaB-alpha(h){p} + Skp1(h) <==> Cul-1(h):beta-TrCP1(m):IkappaB-alpha(h){p}:Skp1(h) (binding) | Cul-1(h) + Cdc25A(h) <==> Cul-1(h):Cdc25A(h) (binding) | beta-TrCP1(h) + Cul-1(h) <==> beta-TrCP1(h):Cul-1(h) (binding) | HOS(h) + Cul-1(h) <==> HOS(h):Cul-1(h) (binding) | Fbw2(h) + Cul-1(h) <==> Fbw2(h):Cul-1(h) (binding) | Fbw4(h) + Cul-1(h) <==> Fbw4(h):Cul-1(h) (binding) | Fbw5(h) + Cul-1(h) <==> Fbw5(h):Cul-1(h) (binding) | Cul-1(h) + Fbw7(h) <==> Cul-1(h):Fbw7(h) (binding) | Cul-1(h) + parkin(h) <==> Cul-1(h):parkin(h) (binding) | Cul-1(h) + 19S proteasome(h) <==> Cul-1(h):19S proteasome(h) (binding) | 20S proteasome(h) + Cul-1(h) <==> 20S proteasome(h):Cul-1(h) (binding) | PSMA2(h) + Cul-1(h) <==> PSMA2(h):Cul-1(h) (binding) | PSMA4(h) + Cul-1(h) <==> PSMA4(h):Cul-1(h) (binding) | PSMA6(h) + Cul-1(h) <==> PSMA6(h):Cul-1(h) (binding) | Cul-1(h) + Fbx33(m) <==> Cul-1(h):Fbx33(m) (binding) | Skp1(h) + Cul-1(h) + Fbx33(m) + YB-1(m.s.) <==> Skp1(h):Cul-1(h):Fbx33(m):YB-1(m.s.) (binding) | Cul-1(h) + Nedd8(h) --APP-BP1(m.s.):Uba3(m.s.),Ubc12(m.s.)--> Cul-1(h){nedd} (neddylation) | Cul-1(h) + Nedd8(h) --APP-BP1(h):Uba3(h),Ubc12(m.s.)--> Cul-1(h){nedd} (neddylation) | Cul-1(h) + Tip120a(h) <==> Tip120a(h):Cul-1(h) (binding) | Roc1(h) + Cul-1(h) + Skp1(h) + Fbx11-isoform1(h) <==> Roc1(h):Cul-1(h):Skp1(h):Fbx11-isoform1(h) (binding) | WWTR1(m) + Cul-1(h) <==> WWTR1(m):Cul-1(h) (binding) | Fbx4(h) + Cul-1(h) <==> Fbx4(h):Cul-1(h) (binding) | reprimo(h) + Cul-1(h) <==> reprimo(h):Cul-1(h) (binding) | T3R-alpha(h) + Cul-1(h) <==> T3R-alpha(h):Cul-1(h) (binding) | REVERB-beta(h) + Cul-1(h) <==> REVERB-beta(h):Cul-1(h) (binding) | Skp2(m.s.){pS72} + Cul-1(h) + Skp1(h) <==> Skp2(m.s.){pS72}:Cul-1(h):Skp1(h) (binding) | Cul-1(h) + Skp1(h) + Skp2(h){pS72} <==> Cul-1(h):Skp1(h):Skp2(h){pS72} (binding) | Cul-1(h) + Skp1(h) + Skp2-alpha(h) <==> Cul-1(h):Skp1(h):Skp2-alpha(h) (binding) | CUL1(h) --> Cul-1(h) (expression). | CUL1(h) --> Cul-1(h) (expression). |
|---|
Post Transcriptional Modification [Top] | |||
|
Location (AA) |
PTM type |
Literature |
Database |
720 | Glycyl lysine isopeptide (Lys-Gly)(interchain with G-Cter in NEDD8). | Swiss-Prot 53.0 |
|---|---|---|---|
Methylation Profile [Top] | ||
|
Chromosome |
Location |
Source |
|---|---|---|
chr7 |
Promoter: 148025366 - 148027366 | Agilent_014791 |
chr7 |
Promoter: 148025366 - 148027366 | GSE27584 |
chr7 |
Promoter: 148025366 - 148027366 | Agilent_014791_44K |
chr7 |
Promoter: 148025366 - 148027366 | Agilent_014791 |