|
||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for PIK3R1 |
| Basic gene info. | Gene symbol | PIK3R1 |
| Gene name | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | |
| Synonyms | AGM7|GRB1|IMD36|p85|p85-ALPHA | |
| Cytomap | UCSC genome browser: 5q13.1 | |
| Genomic location | chr5 :67586466-67597649 | |
| Type of gene | protein-coding | |
| RefGenes | NM_001242466.1, NM_181504.3,NM_181523.2,NM_181524.1, | |
| Ensembl id | ENSG00000145675 | |
| Description | PI3-kinase subunit p85-alphaPI3K regulatory subunit alphaphosphatidylinositol 3-kinase 85 kDa regulatory subunit alphaphosphatidylinositol 3-kinase regulatory subunit alphaphosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha)ph | |
| Modification date | 20141221 | |
| dbXrefs | MIM : 171833 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000145675 | ||
| HPRD : 01381 | ||
| Vega : OTTHUMG00000131251 | ||
| Protein | UniProt: P27986 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_PIK3R1 | |
| BioGPS: 5295 | ||
| Gene Expression Atlas: ENSG00000145675 | ||
| The Human Protein Atlas: ENSG00000145675 | ||
| Pathway | NCI Pathway Interaction Database: PIK3R1 | |
| KEGG: PIK3R1 | ||
| REACTOME: PIK3R1 | ||
| ConsensusPathDB | ||
| Pathway Commons: PIK3R1 | ||
| Metabolism | MetaCyc: PIK3R1 | |
| HUMANCyc: PIK3R1 | ||
| Regulation | Ensembl's Regulation: ENSG00000145675 | |
| miRBase: chr5 :67,586,466-67,597,649 | ||
| TargetScan: NM_001242466 | ||
| cisRED: ENSG00000145675 | ||
| Context | iHOP: PIK3R1 | |
| cancer metabolism search in PubMed: PIK3R1 | ||
| UCL Cancer Institute: PIK3R1 | ||
| Assigned class in ccmGDB | A - This gene has a literature evidence and it belongs to cancer gene. | |
| References showing role of PIK3R1 in cancer cell metabolism | 1. Hellwinkel OJ, Rogmann JP, Asong LE, Luebke AM, Eichelberg C, et al. (2008) A comprehensive analysis of transcript signatures of the phosphatidylinositol‐3 kinase/protein kinase B signal‐transduction pathway in prostate cancer. BJU international 101: 1454-1460. go to article | |
| Top |
| Phenotypic Information for PIK3R1(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: PIK3R1 |
| Familial Cancer Database: PIK3R1 | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
| . | ||||||||||||||
Oncogene 1 | ||||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 6 http://www.nature.com/nature/journal/v490/n7418/full/nature11412.html, 7 http://www.sciencedirect.com/science/article/pii/S0092867413012087, 8 http://www.nature.com/nature/journal/v497/n7447/full/nature12113.html, 9 http://www.nejm.org/doi/full/10.1056/NEJMoa1402121, 10Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| KEGG_FC_GAMMA_R_MEDIATED_PHAGOCYTOSIS REACTOME_PHOSPHOLIPID_METABOLISM REACTOME_PI_METABOLISM REACTOME_METABOLISM_OF_LIPIDS_AND_LIPOPROTEINS | |
| OMIM | 171833; gene. 171833; gene. 269880; phenotype. 269880; phenotype. 615214; phenotype. 615214; phenotype. |
| Orphanet | 3163; SHORT syndrome. 3163; SHORT syndrome. 33110; Autosomal agammaglobulinemia. 33110; Autosomal agammaglobulinemia. |
| Disease | KEGG Disease: PIK3R1 |
| MedGen: PIK3R1 (Human Medical Genetics with Condition) | |
| ClinVar: PIK3R1 | |
| Phenotype | MGI: PIK3R1 (International Mouse Phenotyping Consortium) |
| PhenomicDB: PIK3R1 | |
| Mutations for PIK3R1 |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| - Statistics for Tissue and Mutation type | Top |
 |
| - For Inter-chromosomal Variations |
| * Inter-chromosomal variantions includes 'interchromosomal amplicon to amplicon', 'interchromosomal amplicon to non-amplified dna', 'interchromosomal insertion', 'Interchromosomal unknown type'. |
 |
| - For Intra-chromosomal Variations |
| * Intra-chromosomal variantions includes 'intrachromosomal amplicon to amplicon', 'intrachromosomal amplicon to non-amplified dna', 'intrachromosomal deletion', 'intrachromosomal fold-back inversion', 'intrachromosomal inversion', 'intrachromosomal tandem duplication', 'Intrachromosomal unknown type', 'intrachromosomal with inverted orientation', 'intrachromosomal with non-inverted orientation'. |
 |
| Sample | Symbol_a | Chr_a | Start_a | End_a | Symbol_b | Chr_b | Start_b | End_b |
| breast | PIK3R1 | chr5 | 67587611 | 67587611 | chr16 | 74740421 | 74740421 | |
| central_nervous_system | PIK3R1 | chr5 | 67590608 | 67590608 | chr5 | 67465996 | 67465996 |
| cf) Tissue number; Tissue name (1;Breast, 2;Central_nervous_system, 3;Haematopoietic_and_lymphoid_tissue, 4;Large_intestine, 5;Liver, 6;Lung, 7;Ovary, 8;Pancreas, 9;Prostate, 10;Skin, 11;Soft_tissue, 12;Upper_aerodigestive_tract) |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows PIK3R1 related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| BF849250 | LRCH1 | 6 | 162 | 13 | 47155365 | 47155523 | PIK3R1 | 144 | 440 | 5 | 67593268 | 67593562 | |
| BX648791 | NFIA | 1 | 4560 | 1 | 61817423 | 61821979 | PIK3R1 | 4560 | 5386 | 5 | 67593366 | 67594192 | |
| BU742563 | PIK3R1 | 8 | 278 | 5 | 67594299 | 67594570 | PIK3R1 | 272 | 651 | 5 | 67595900 | 67596280 | |
| Top |
| There's no copy number variation information in COSMIC data for this gene. |
| Top |
|
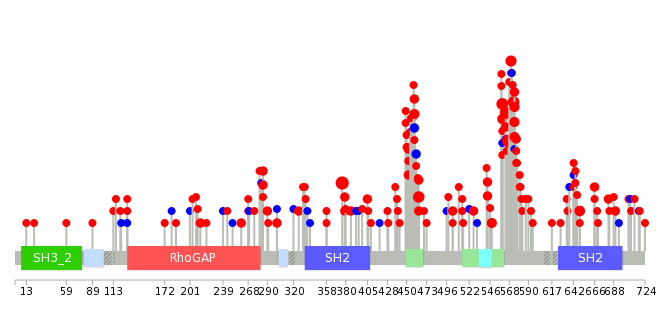 |
| Top |
| Stat. for Non-Synonymous SNVs (# total SNVs=251) | (# total SNVs=27) |
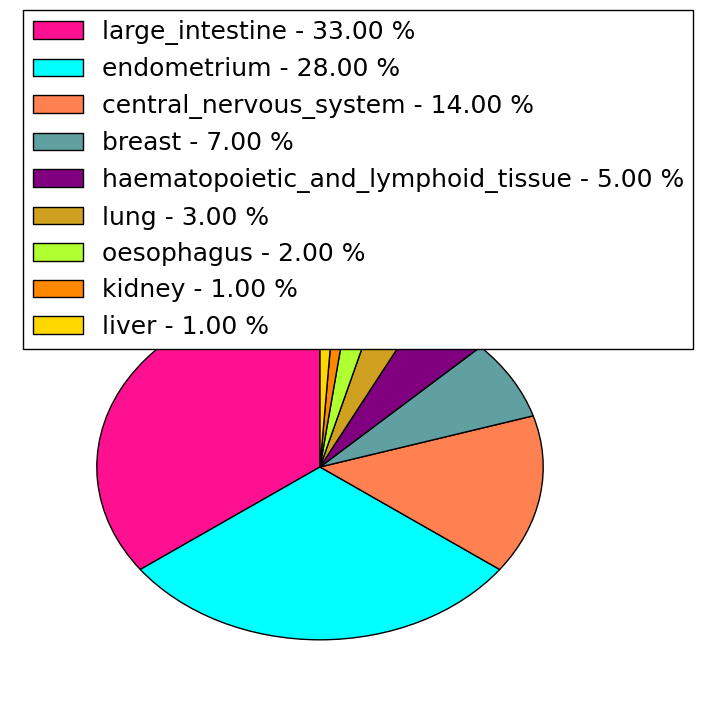 | 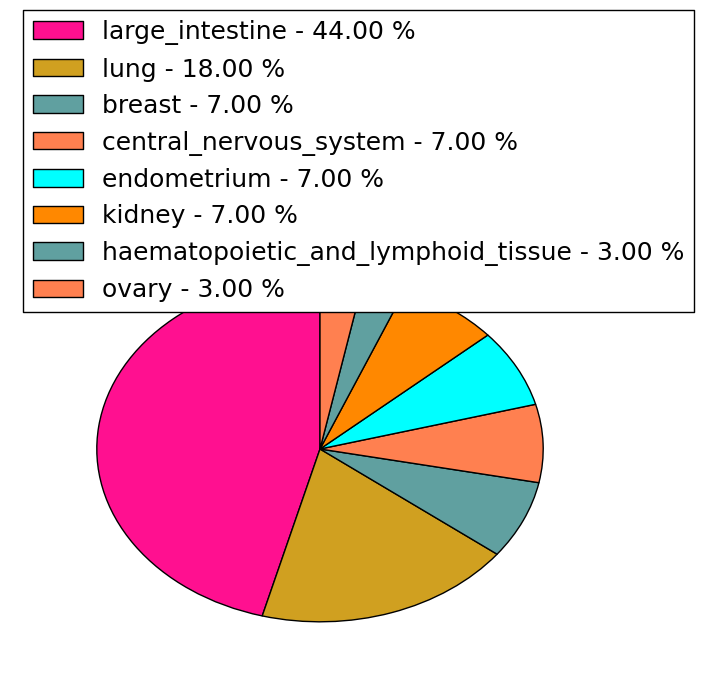 |
(# total SNVs=97) | (# total SNVs=31) |
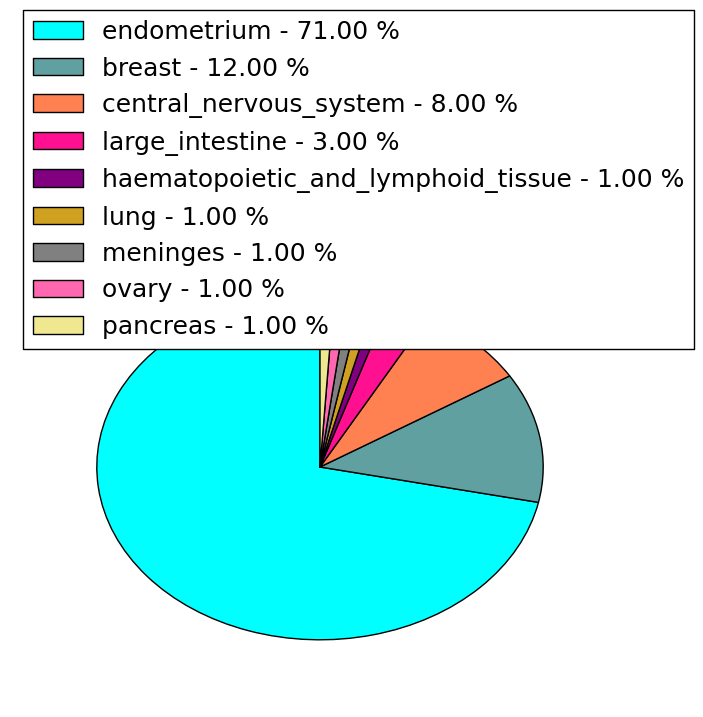 | 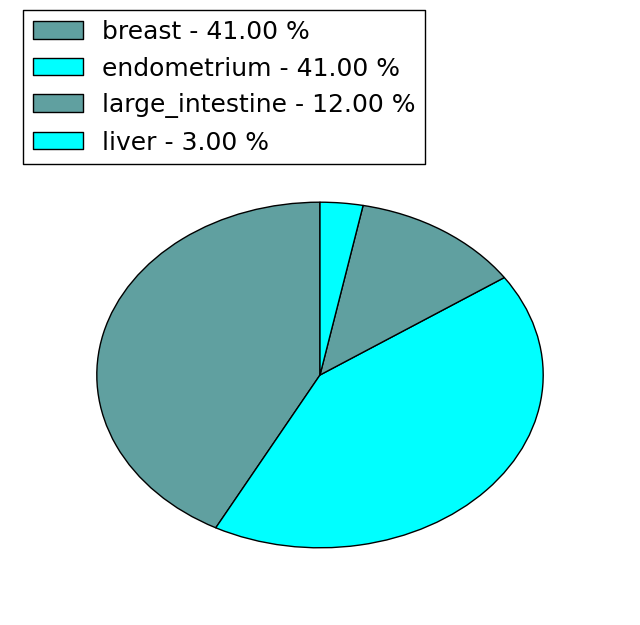 |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr5:67588951-67588951 | p.R348* | 22 |
| chr5:67589138-67589138 | p.G376R | 12 |
| chr5:67591106-67591106 | p.K567E | 8 |
| chr5:67591086-67591086 | p.D560G | 7 |
| chr5:67591097-67591097 | p.N564D | 7 |
| chr5:67591246-67591246 | p.? | 7 |
| chr5:67591125-67591125 | p.L573P | 6 |
| chr5:67589628-67589628 | p.D464G | 6 |
| chr5:67591128-67591128 | p.R574I | 6 |
| chr5:67592108-67592108 | p.R642* | 6 |
| Top |
|
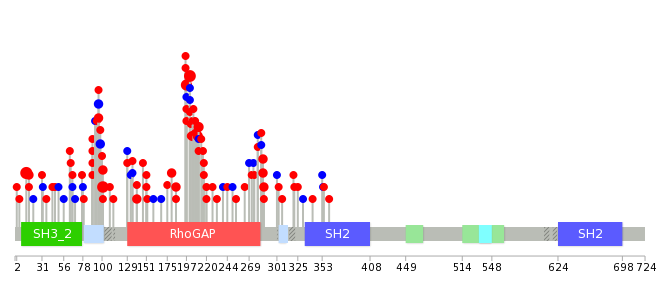 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 2 | 9 | 1 | 48 | 16 | 4 | 2 | 2 | 6 | 3 | 2 | 6 | 6 | 1 | 27 | |||||
| # mutation | 2 | 9 | 1 | 35 | 11 | 4 | 2 | 3 | 6 | 3 | 2 | 6 | 6 | 1 | 28 | |||||
| nonsynonymous SNV | 2 | 8 | 1 | 24 | 10 | 4 | 2 | 1 | 2 | 2 | 1 | 2 | 6 | 1 | 25 | |||||
| synonymous SNV | 1 | 11 | 1 | 2 | 4 | 1 | 1 | 4 | 3 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr5:67589138 | p.G13R,PIK3R1 | 8 |
| chr5:67589628 | p.D101G,PIK3R1 | 7 |
| chr5:67591097 | p.N201D,PIK3R1 | 7 |
| chr5:67591086 | p.D197G,PIK3R1 | 5 |
| chr5:67591106 | p.K204E,PIK3R1 | 3 |
| chr5:67590446 | p.L210P,PIK3R1 | 3 |
| chr5:67591125 | p.R211G,PIK3R1 | 3 |
| chr5:67591127 | p.R140L,PIK3R1 | 3 |
| chr5:67591099 | p.Y89N,PIK3R1 | 2 |
| chr5:67576772 | p.K96E,PIK3R1 | 2 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
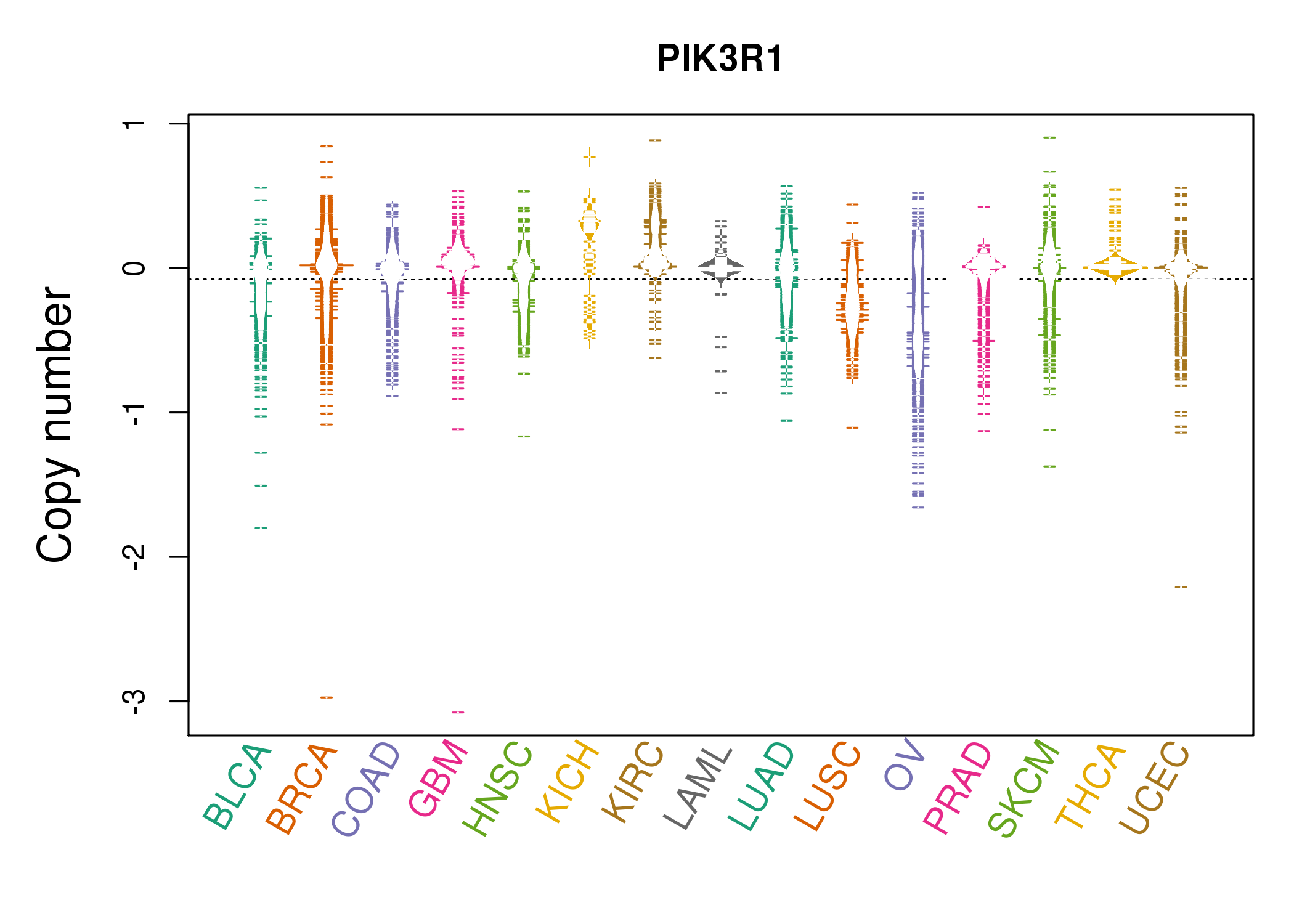 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
| Gene Expression for PIK3R1 |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
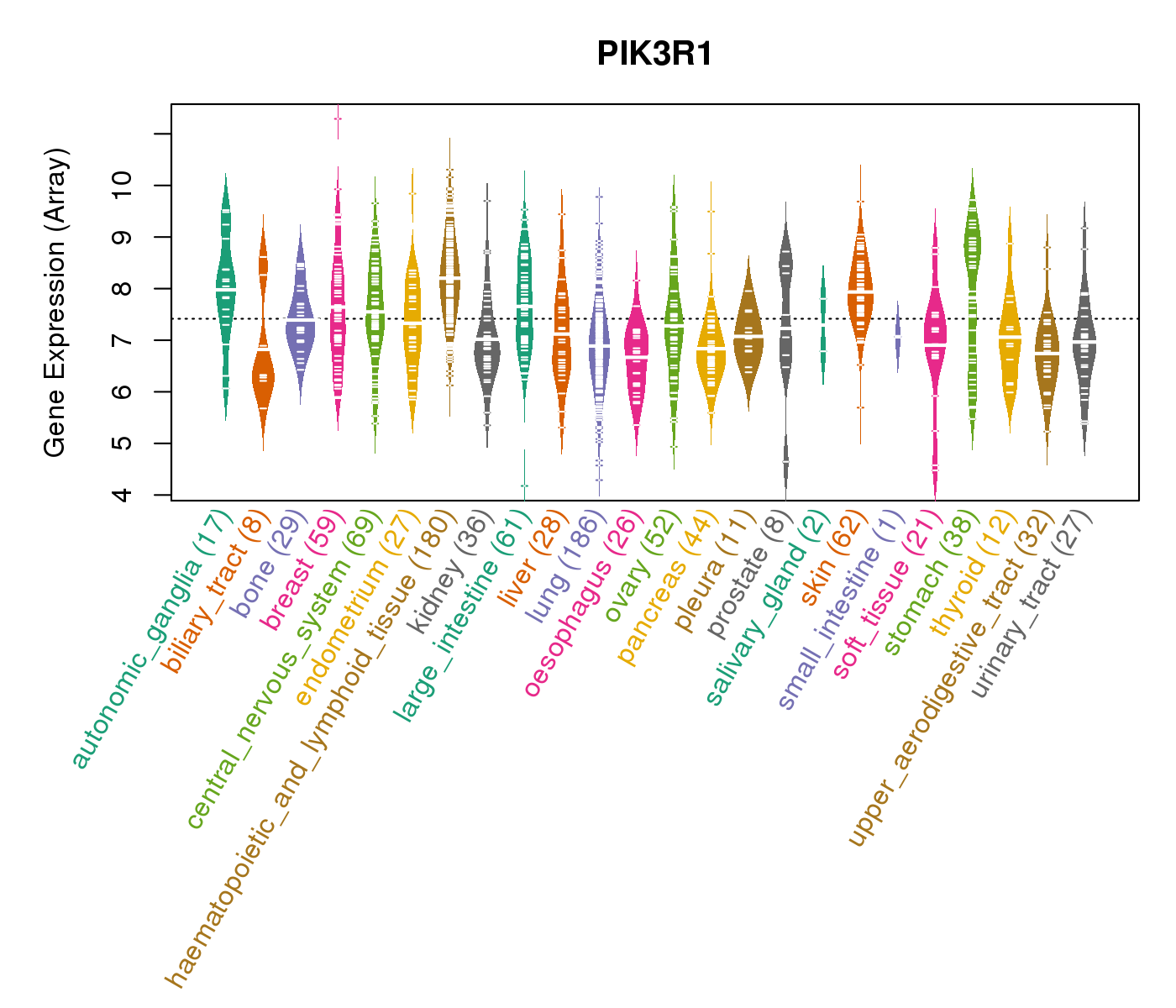 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
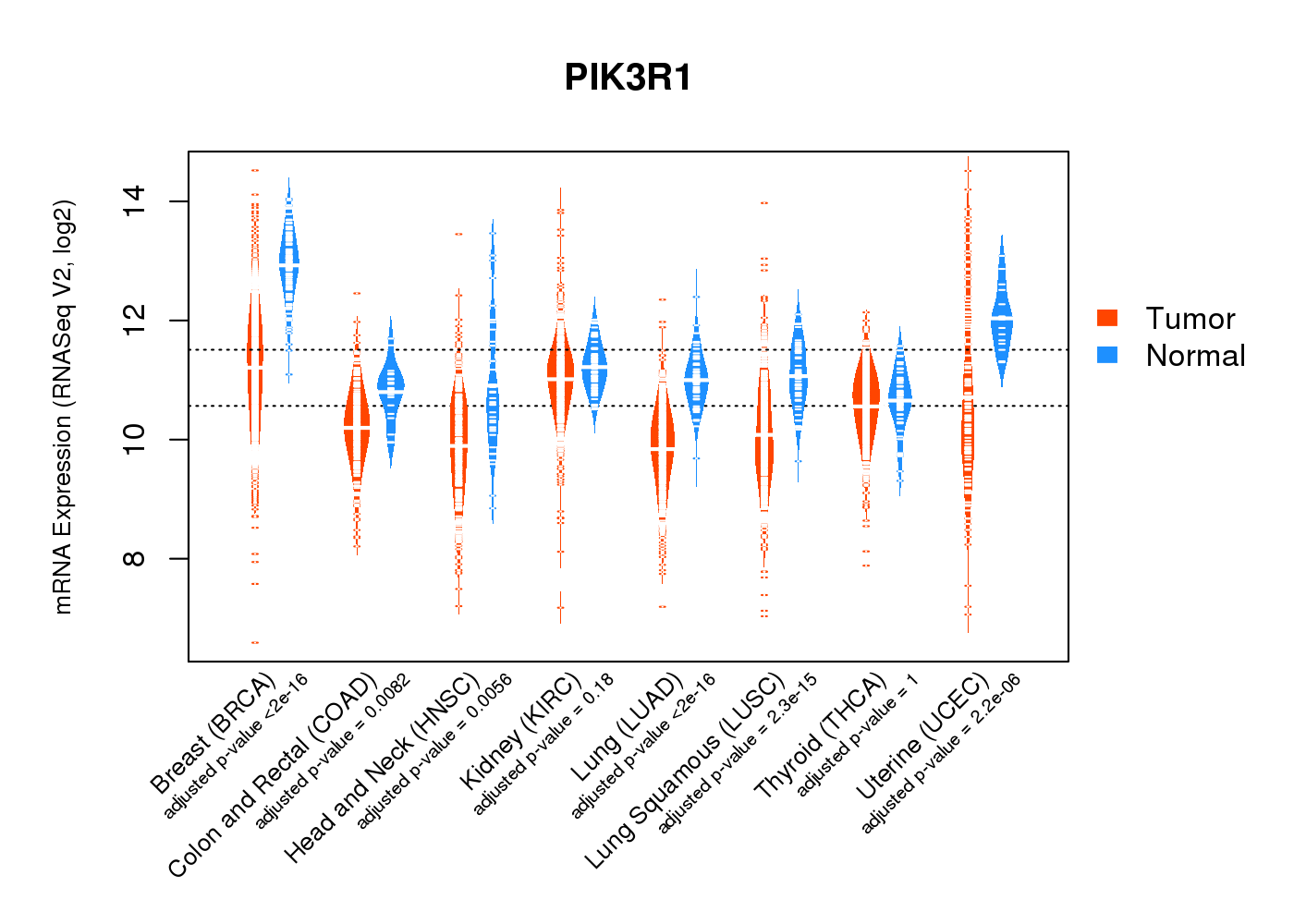 |
| Top |
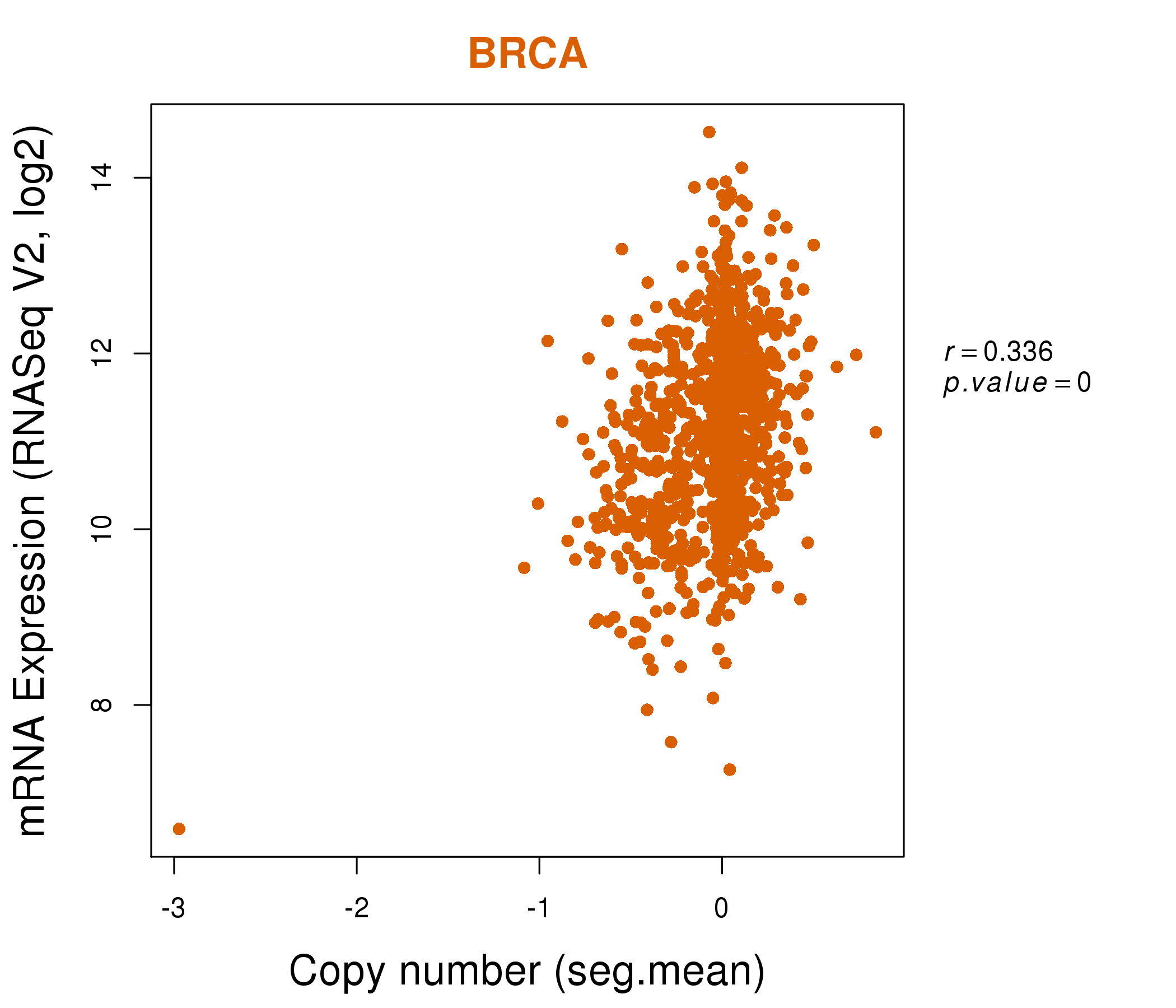
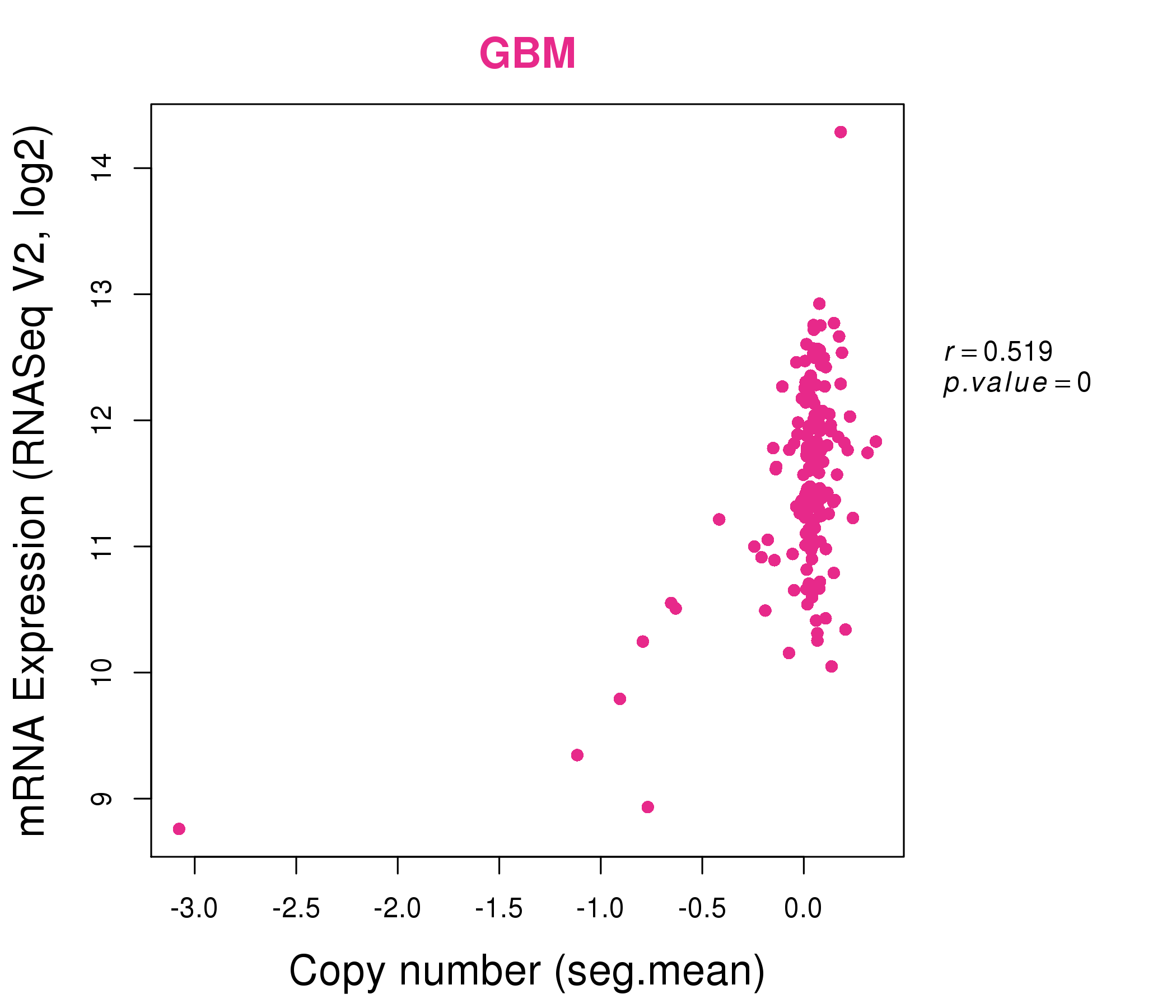
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
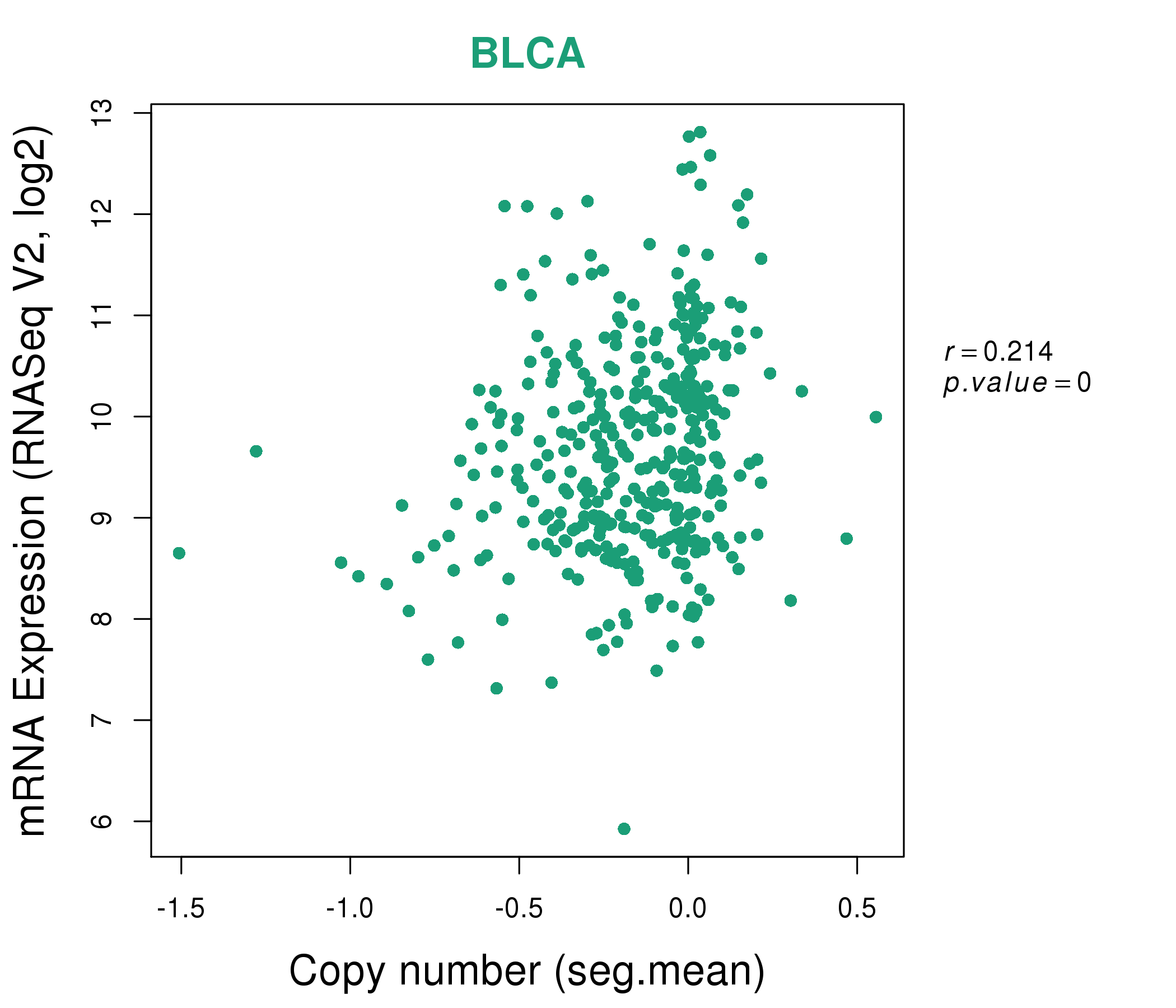 |
|
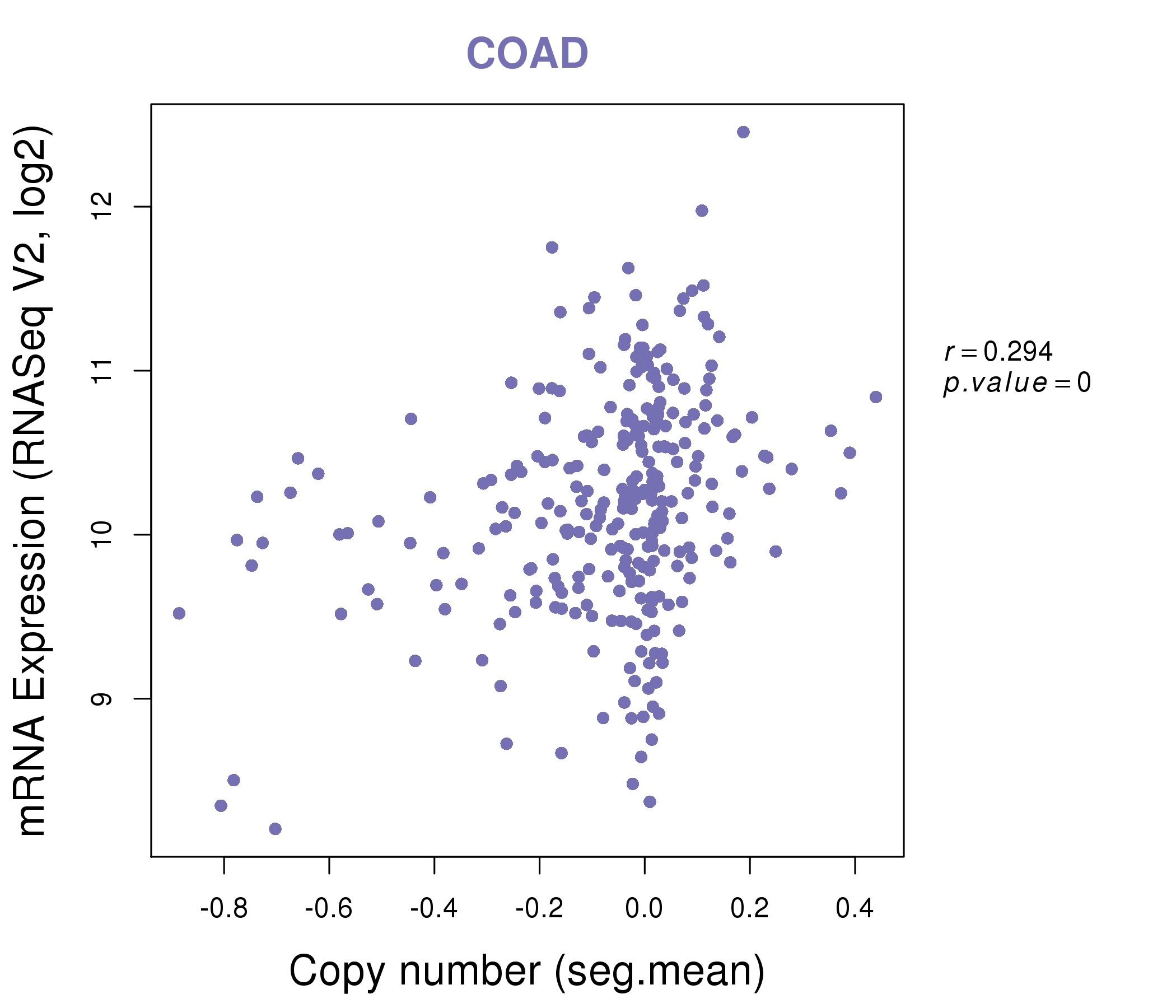 |
|
| Top |
| Gene-Gene Network Information |
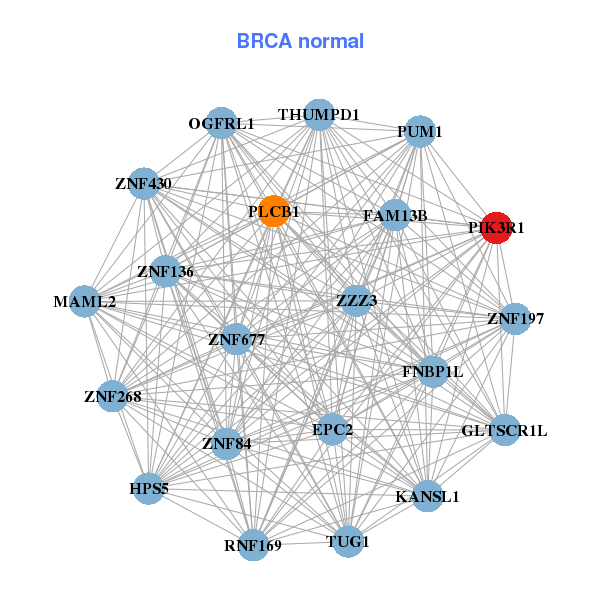
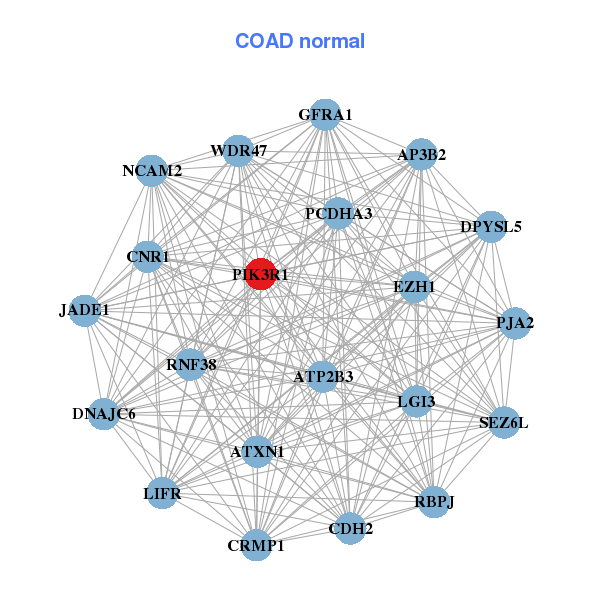
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
 |
| ||||
| ARID5B,ARRDC3,ATF7,MIR99AHG,DST,FRMD4B,IQGAP2, MTUS1,NUMB,PDZD2,PIK3R1,PJA2,PRDM2,RBMS3, RFX7,SH3RF2,SP1,TLR3,TSHZ2,UTRN,YAP1 | EPC2,FAM13B,FNBP1L,HPS5,GLTSCR1L,KANSL1,MAML2, OGFRL1,PIK3R1,PLCB1,PUM1,RNF169,THUMPD1,TUG1, ZNF136,ZNF197,ZNF268,ZNF430,ZNF677,ZNF84,ZZZ3 | ||||
 |
| ||||
| ARL15,ATAD2B,DNMT3A,FOXO3B,GDF10,GOLIM4,KCTD16, KRT32,KRT38,MYL3,MYO10,NAV2,NCKAP5,NRXN3, OLFML3,PHYHIPL,PIK3R1,PRSS23,STK38L,TP53BP1,UGT2A3 | AP3B2,ATP2B3,ATXN1,CDH2,CNR1,CRMP1,DNAJC6, DPYSL5,EZH1,GFRA1,LGI3,LIFR,NCAM2,PCDHA3, JADE1,PIK3R1,PJA2,RBPJ,RNF38,SEZ6L,WDR47 |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for PIK3R1 |
| DB Category | DB Name | DB's ID and Url link |
| Chemistry | BindingDB | P27986; -. |
| Chemistry | ChEMBL | CHEMBL2111367; -. |
| Chemistry | BindingDB | P27986; -. |
| Chemistry | ChEMBL | CHEMBL2111367; -. |
| Organism-specific databases | PharmGKB | PA33312; -. |
| Organism-specific databases | PharmGKB | PA33312; -. |
| Organism-specific databases | CTD | 5295; -. |
| Organism-specific databases | CTD | 5295; -. |
| * Gene Centered Interaction Network. |
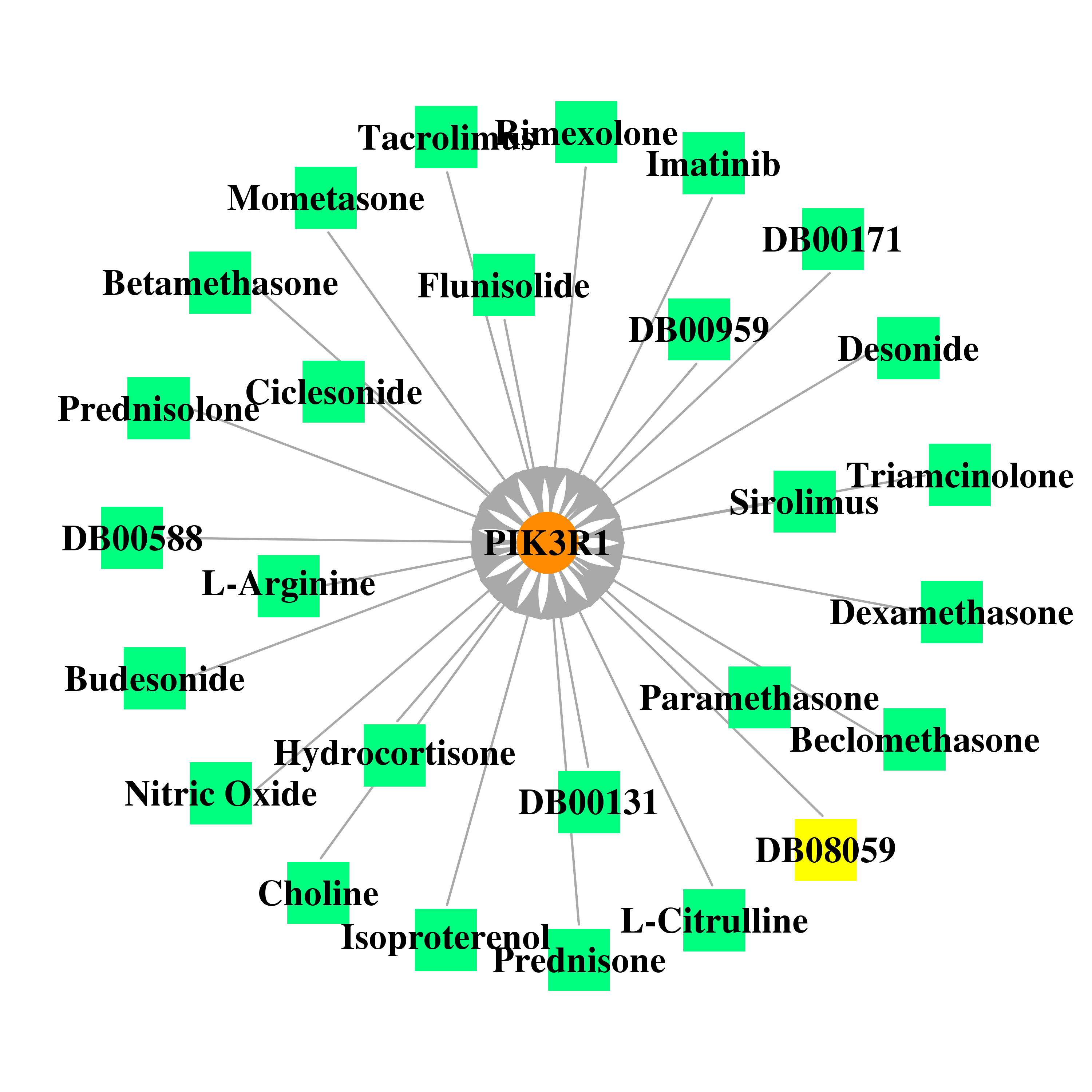 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB01064 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Isoproterenol | 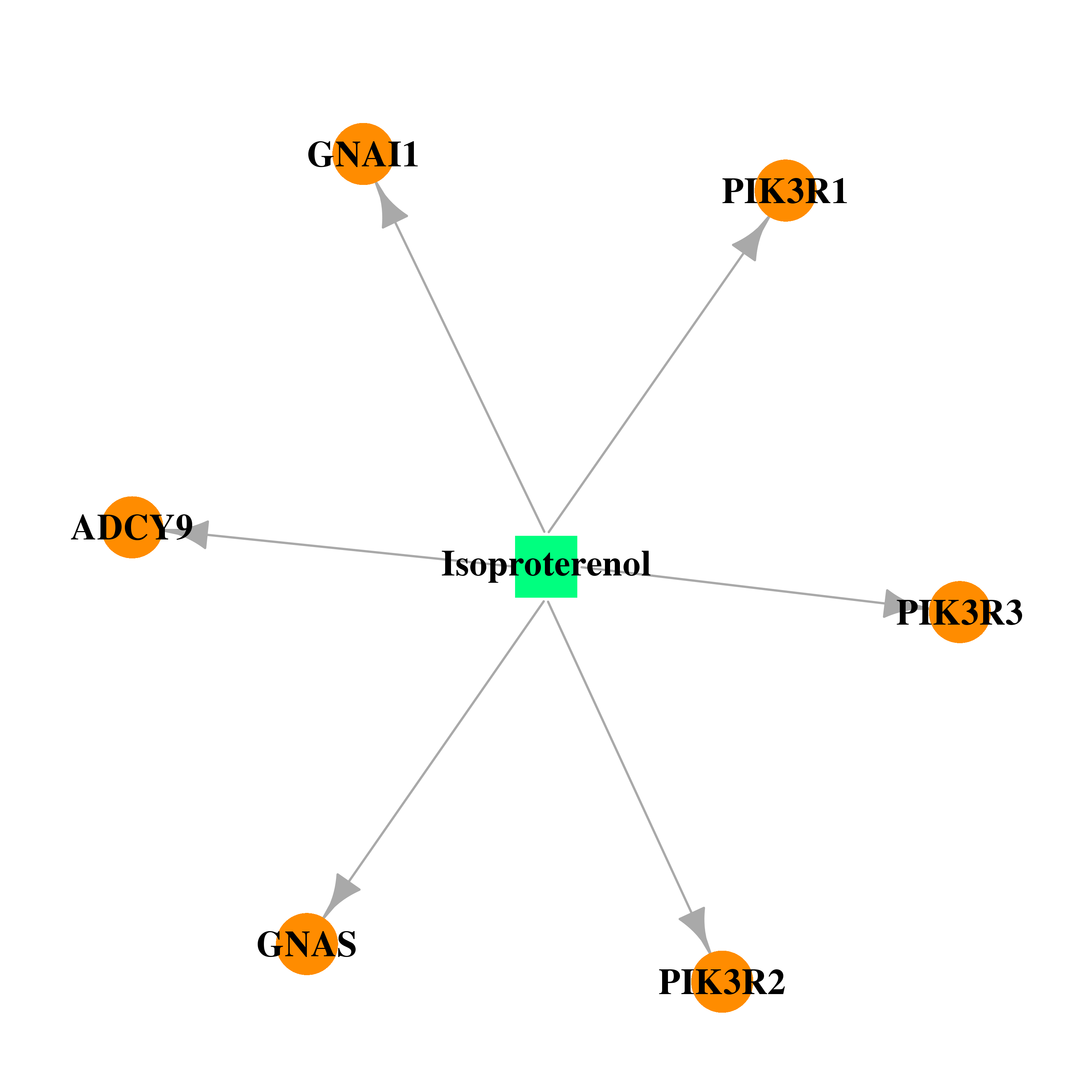 |  |
| DB08059 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | experimental | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4,3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE |  |  |
| DB00171 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; nutraceutical | Adenosine triphosphate |  |  |
| DB00877 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; investigational | Sirolimus |  |  |
| DB00122 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; nutraceutical | Choline |  | 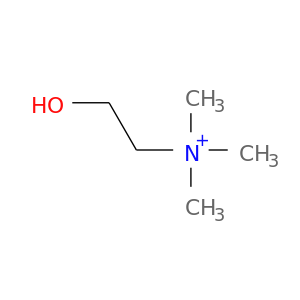 |
| DB00394 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Beclomethasone | 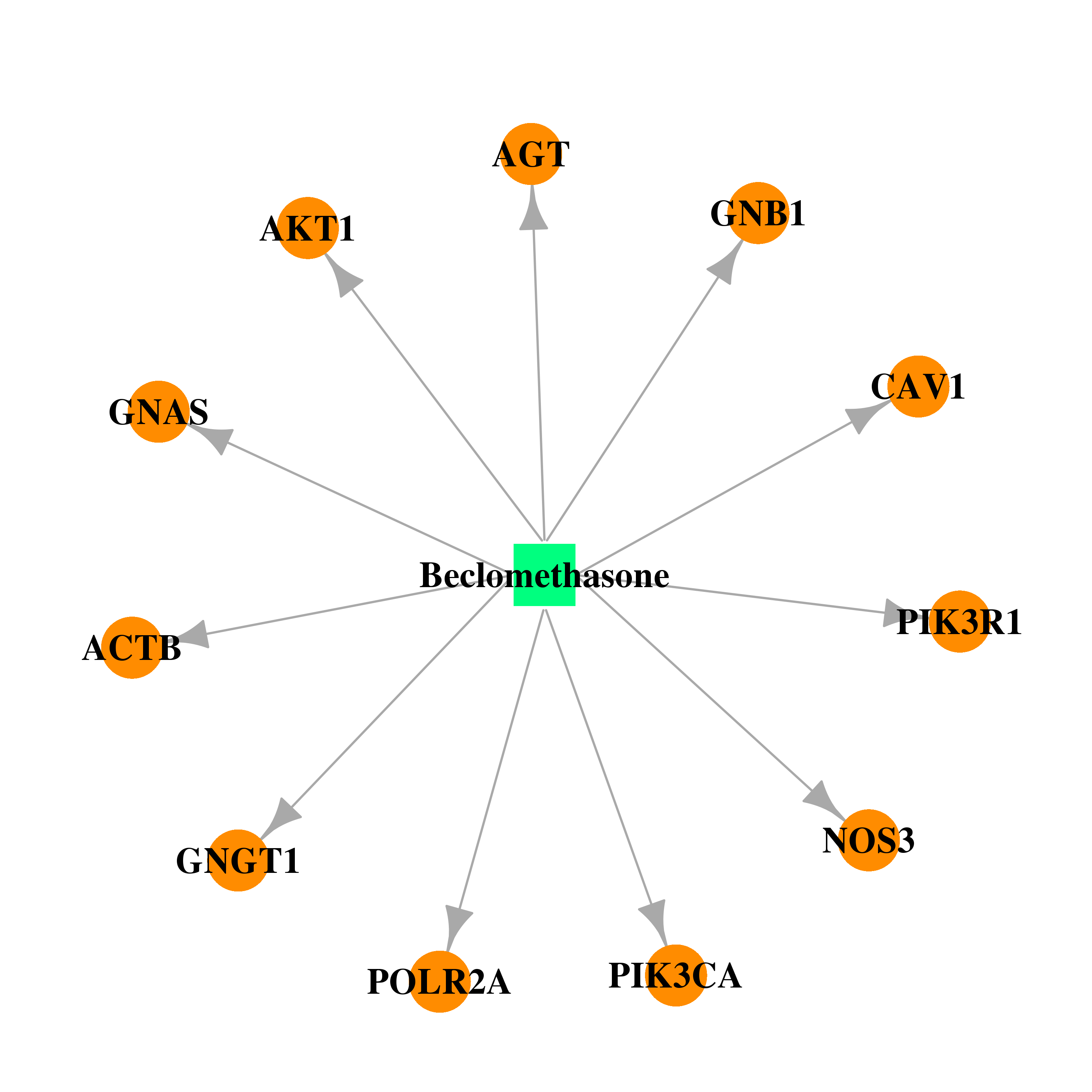 | 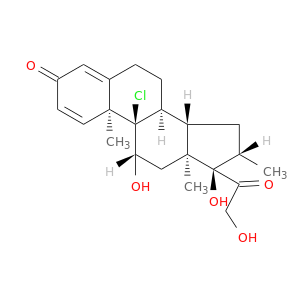 |
| DB00443 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Betamethasone |  | 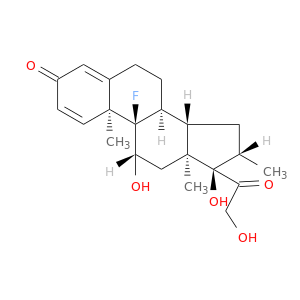 |
| DB01222 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; investigational | Budesonide |  |  |
| DB01410 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; investigational | Ciclesonide |  |  |
| DB01260 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; investigational | Desonide |  |  |
| DB01234 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; investigational | Dexamethasone |  |  |
| DB00180 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; investigational | Flunisolide | 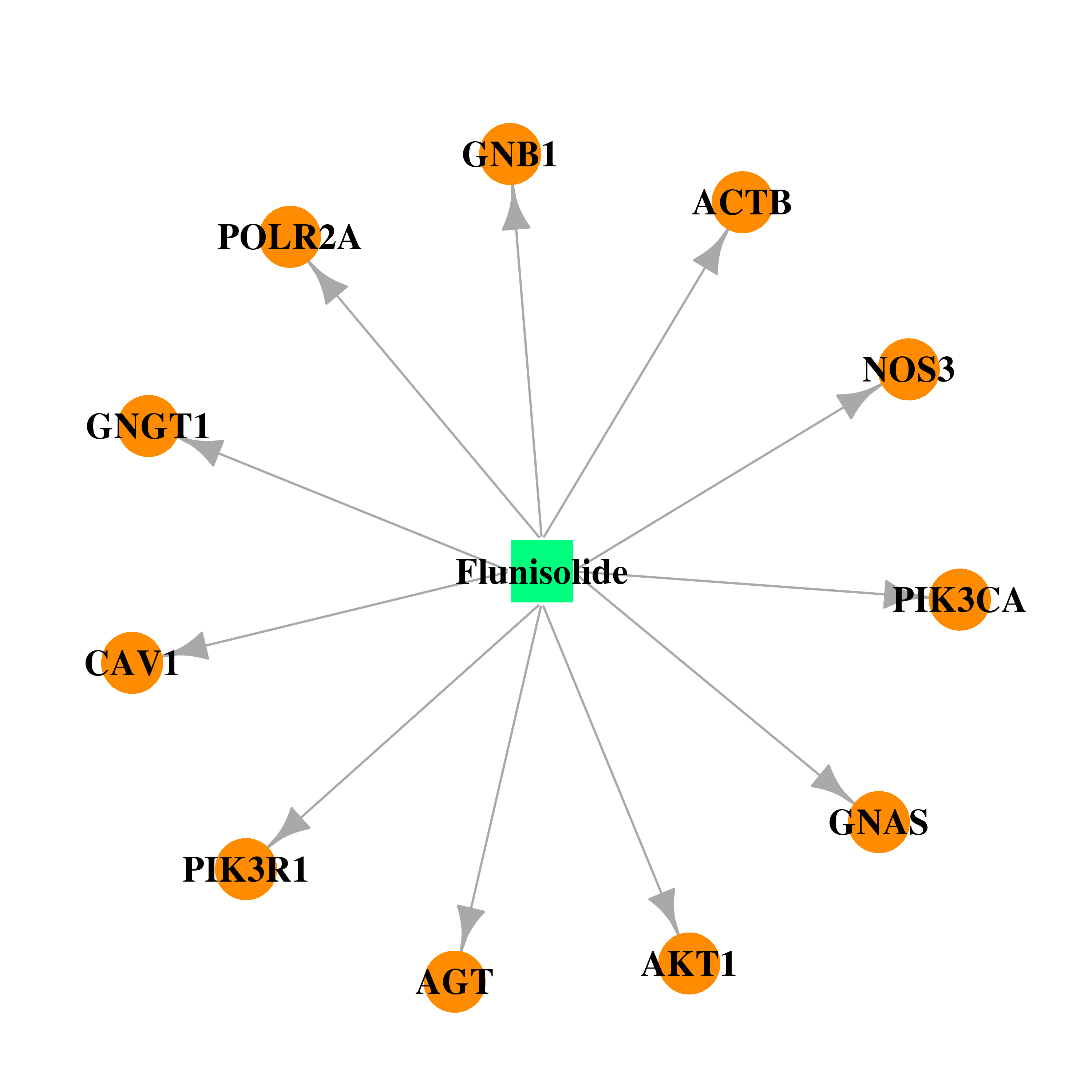 |  |
| DB00588 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; investigational | Fluticasone Propionate | 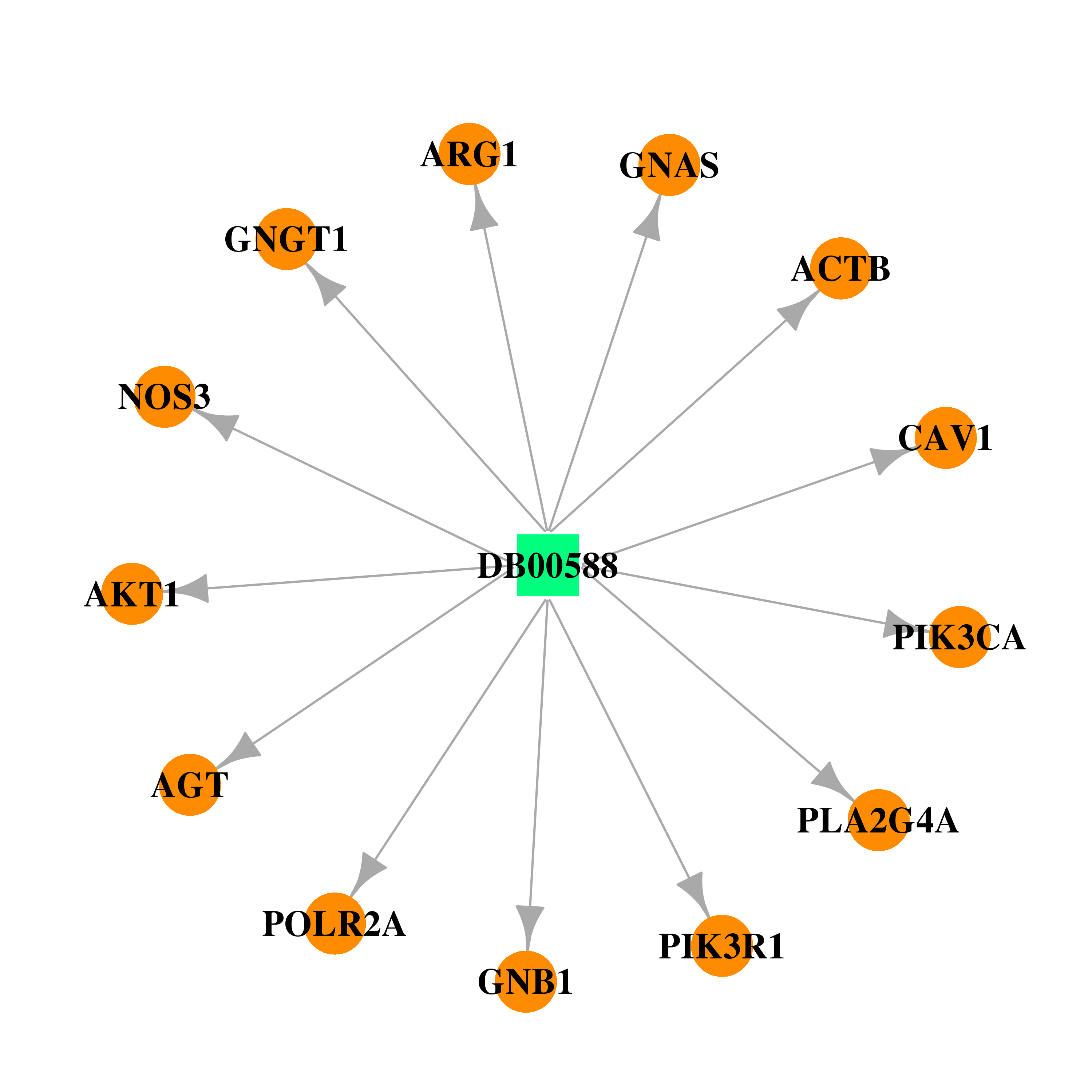 |  |
| DB00741 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Hydrocortisone |  |  |
| DB00959 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Methylprednisolone |  |  |
| DB00764 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Mometasone |  |  |
| DB01384 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Paramethasone |  |  |
| DB00860 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Prednisolone |  |  |
| DB00635 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Prednisone |  |  |
| DB00896 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Rimexolone |  |  |
| DB00620 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Triamcinolone |  |  |
| DB00131 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; nutraceutical | Adenosine monophosphate | 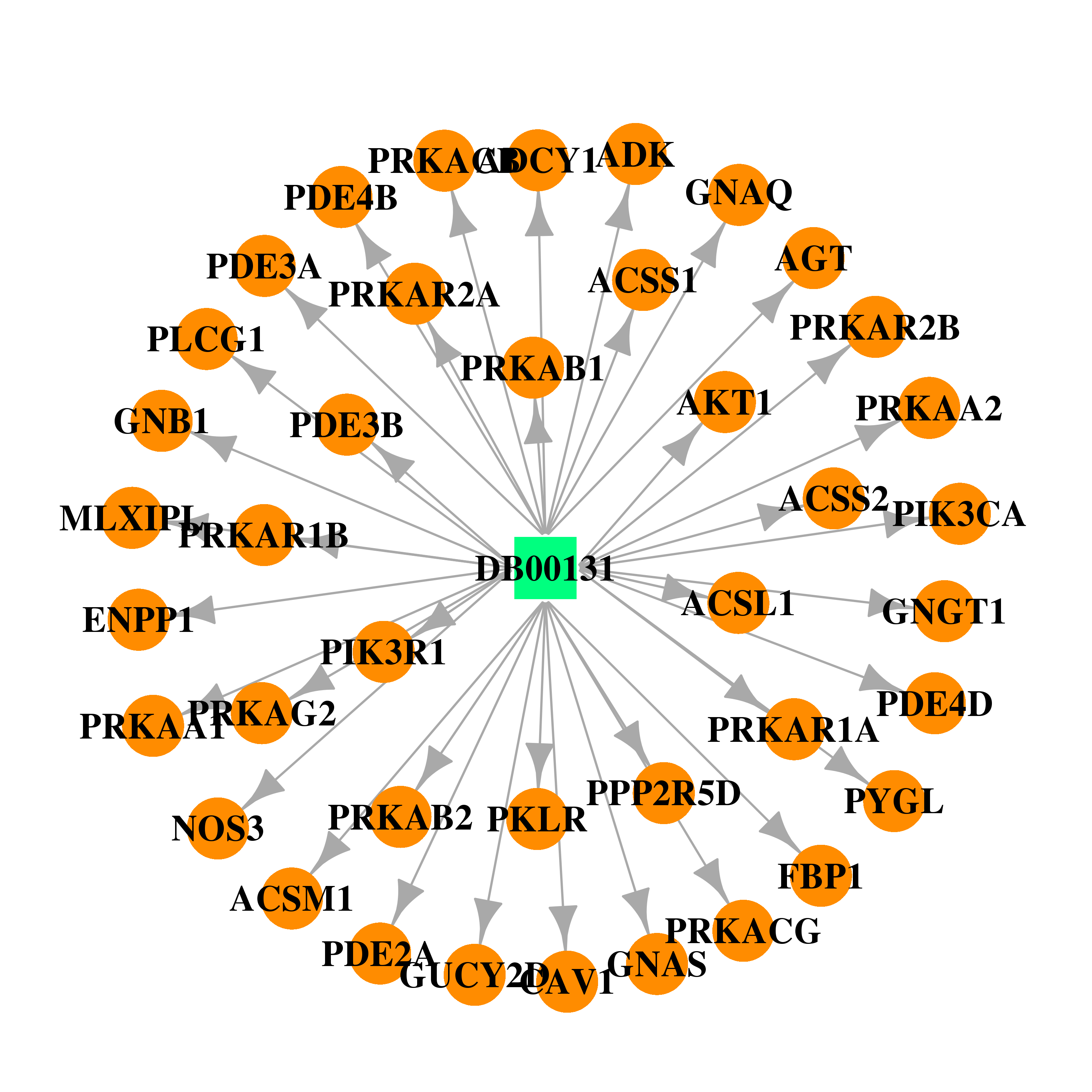 |  |
| DB00125 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; nutraceutical | L-Arginine | 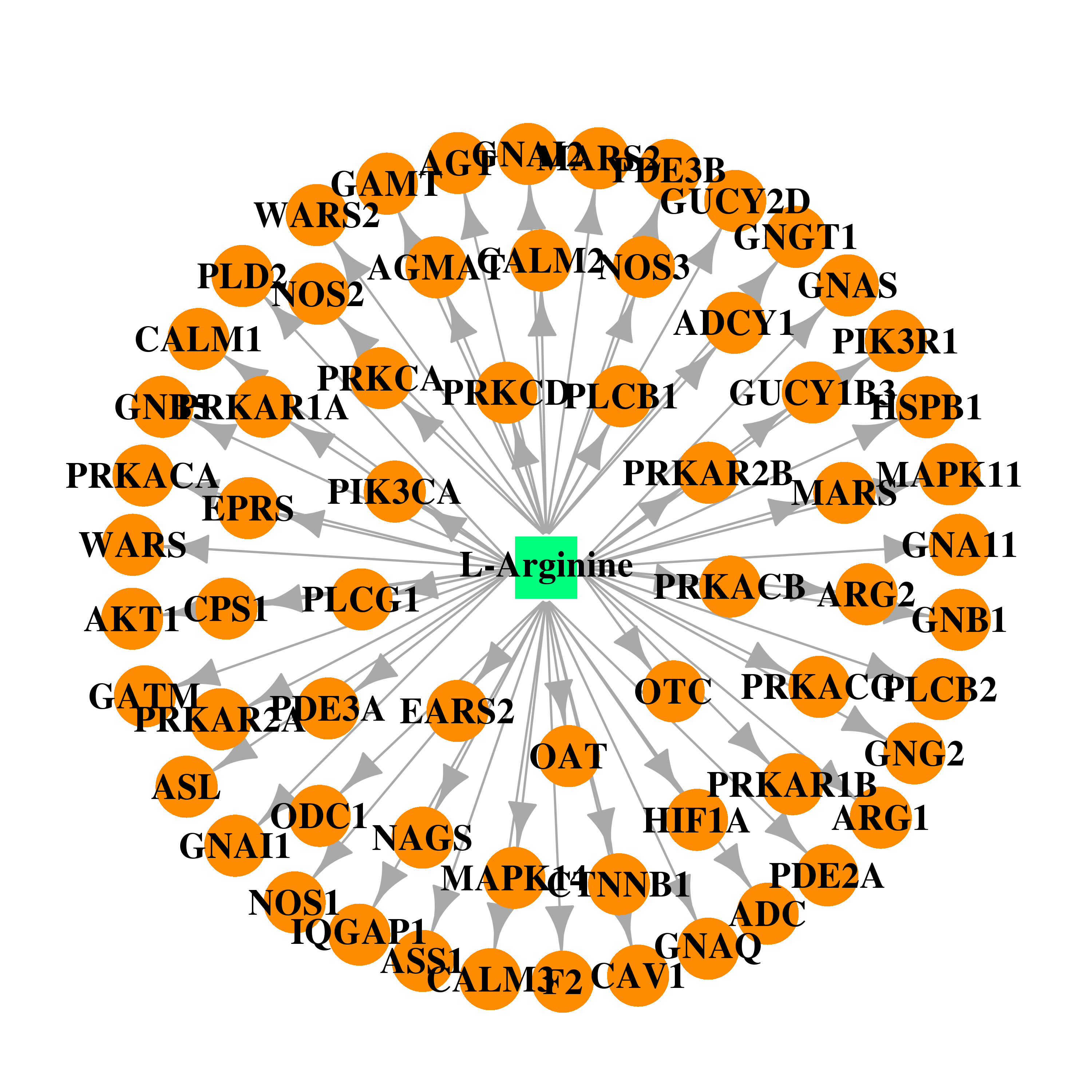 | 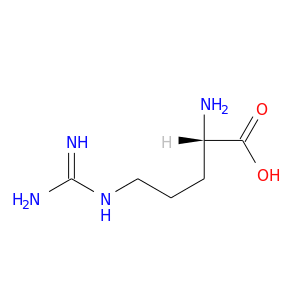 |
| DB00155 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; nutraceutical | L-Citrulline | 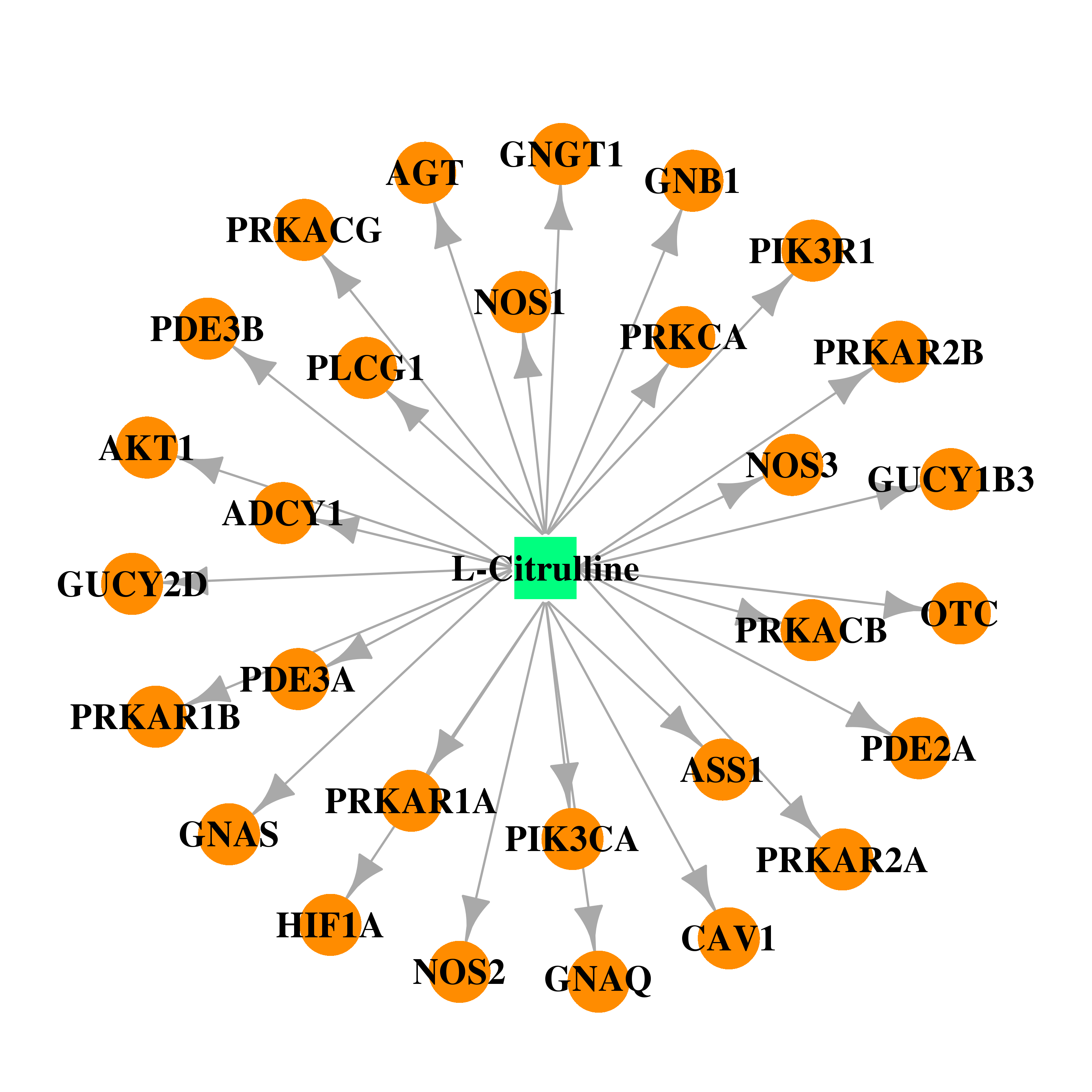 | 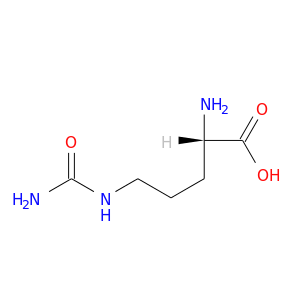 |
| DB00435 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Nitric Oxide | 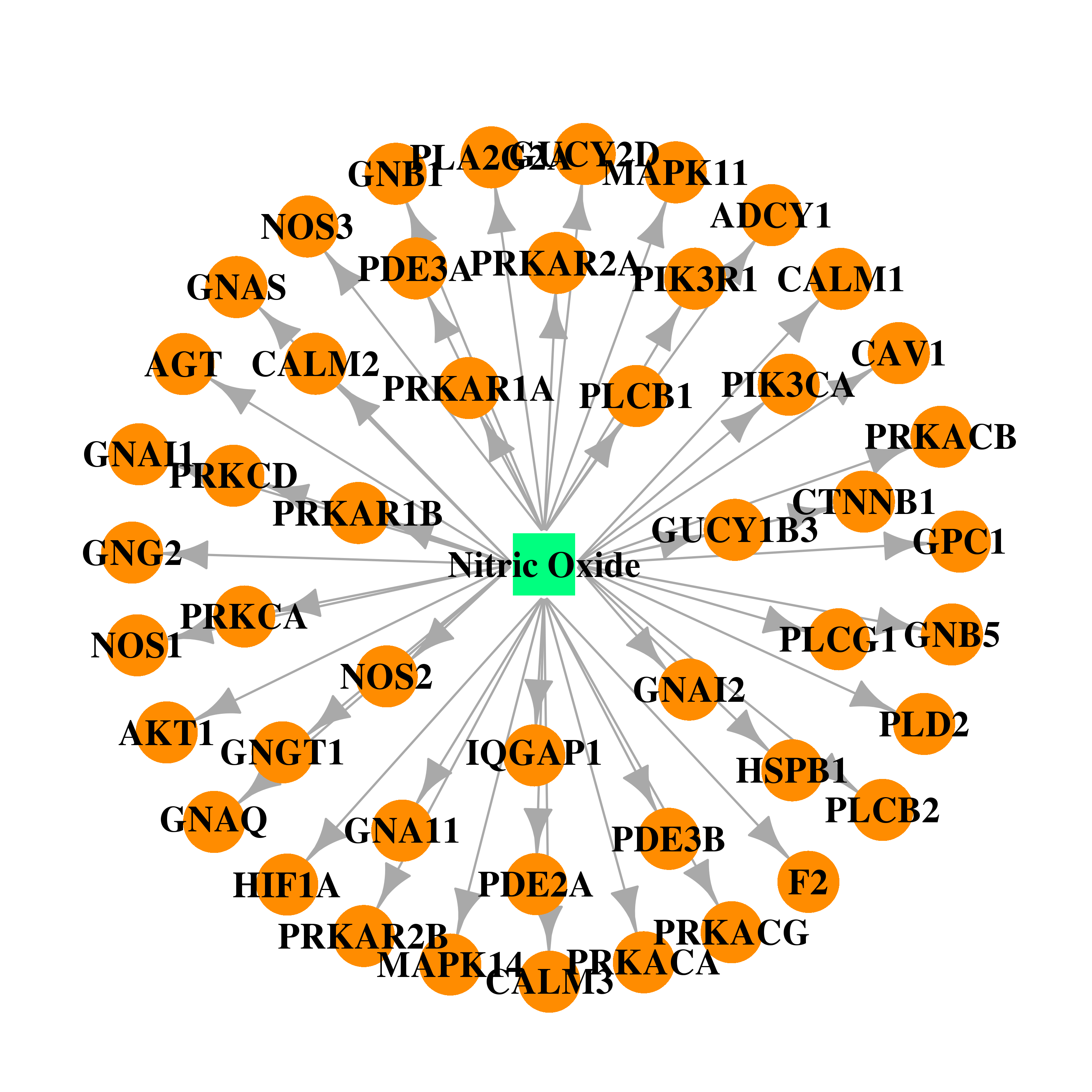 |  |
| DB00864 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved; investigational | Tacrolimus |  |  |
| DB00619 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | approved | Imatinib | 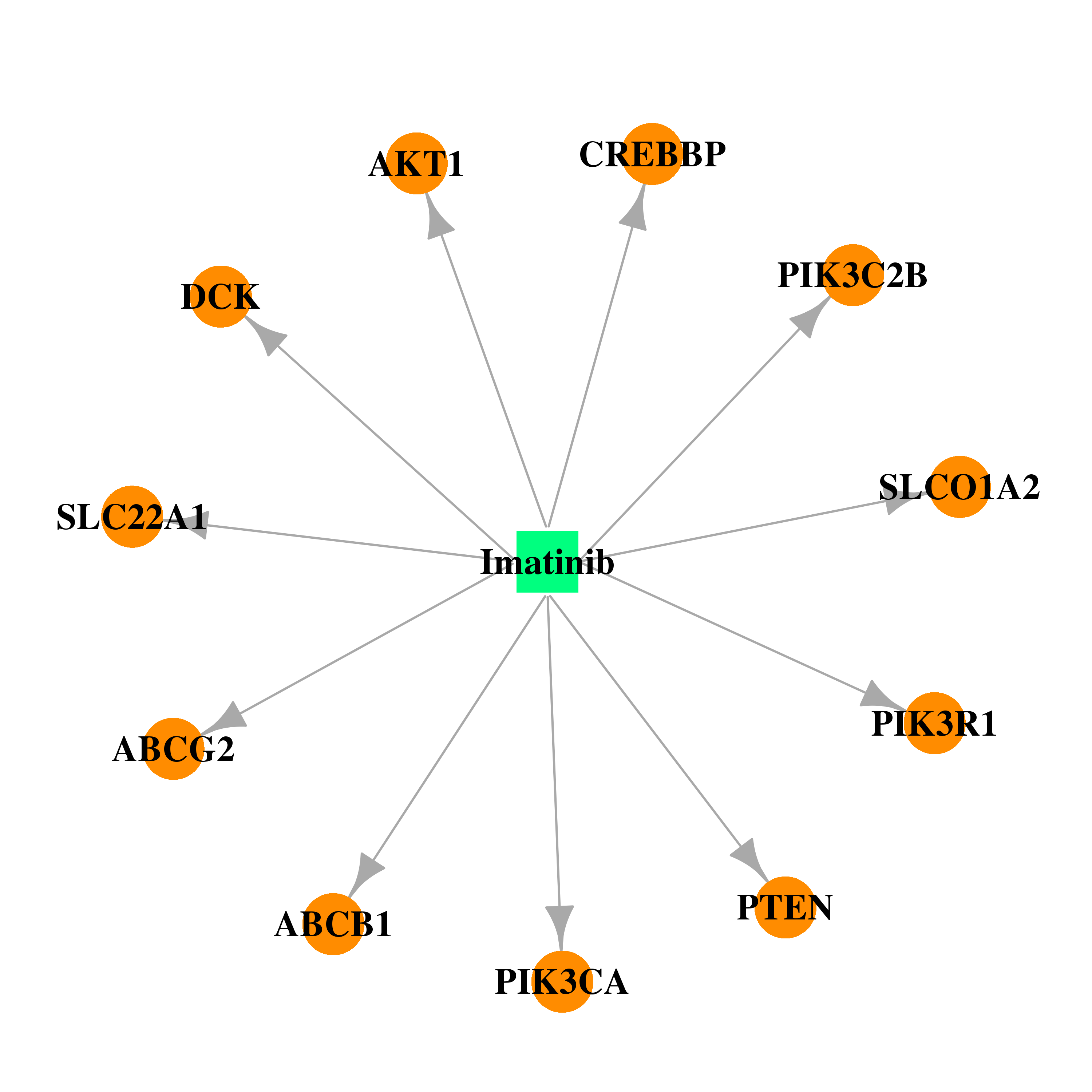 |  |
| Top |
| Cross referenced IDs for PIK3R1 |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |