|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for POLR2A |
| Basic gene info. | Gene symbol | POLR2A |
| Gene name | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | |
| Synonyms | POLR2|POLRA|RPB1|RPBh1|RPO2|RPOL2|RpIILS|hRPB220|hsRPB1 | |
| Cytomap | UCSC genome browser: 17p13.1 | |
| Genomic location | chr17 :7387697-7417935 | |
| Type of gene | protein-coding | |
| RefGenes | NM_000937.4, | |
| Ensembl id | ENSG00000181222 | |
| Description | DNA-directed RNA polymerase II largest subunit, RNA polymerase II 220 kd subunitDNA-directed RNA polymerase II subunit ADNA-directed RNA polymerase II subunit RPB1DNA-directed RNA polymerase III largest subunitRNA polymerase II subunit B1RNA-directed | |
| Modification date | 20141207 | |
| dbXrefs | MIM : 180660 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000181222 | ||
| HPRD : 08916 | ||
| Vega : OTTHUMG00000177594 | ||
| Protein | UniProt: P24928 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_POLR2A | |
| BioGPS: 5430 | ||
| Gene Expression Atlas: ENSG00000181222 | ||
| The Human Protein Atlas: ENSG00000181222 | ||
| Pathway | NCI Pathway Interaction Database: POLR2A | |
| KEGG: POLR2A | ||
| REACTOME: POLR2A | ||
| ConsensusPathDB | ||
| Pathway Commons: POLR2A | ||
| Metabolism | MetaCyc: POLR2A | |
| HUMANCyc: POLR2A | ||
| Regulation | Ensembl's Regulation: ENSG00000181222 | |
| miRBase: chr17 :7,387,697-7,417,935 | ||
| TargetScan: NM_000937 | ||
| cisRED: ENSG00000181222 | ||
| Context | iHOP: POLR2A | |
| cancer metabolism search in PubMed: POLR2A | ||
| UCL Cancer Institute: POLR2A | ||
| Assigned class in ccmGDB | B - This gene belongs to cancer gene. | |
| Top |
| Phenotypic Information for POLR2A(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: POLR2A |
| Familial Cancer Database: POLR2A | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
| . | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Oncogene 1 | Significant driver gene in | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| KEGG_PURINE_METABOLISM KEGG_PYRIMIDINE_METABOLISM | |
| OMIM | 180660; gene+phenotype. |
| Orphanet | |
| Disease | KEGG Disease: POLR2A |
| MedGen: POLR2A (Human Medical Genetics with Condition) | |
| ClinVar: POLR2A | |
| Phenotype | MGI: POLR2A (International Mouse Phenotyping Consortium) |
| PhenomicDB: POLR2A | |
| Mutations for POLR2A |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| - Statistics for Tissue and Mutation type | Top |
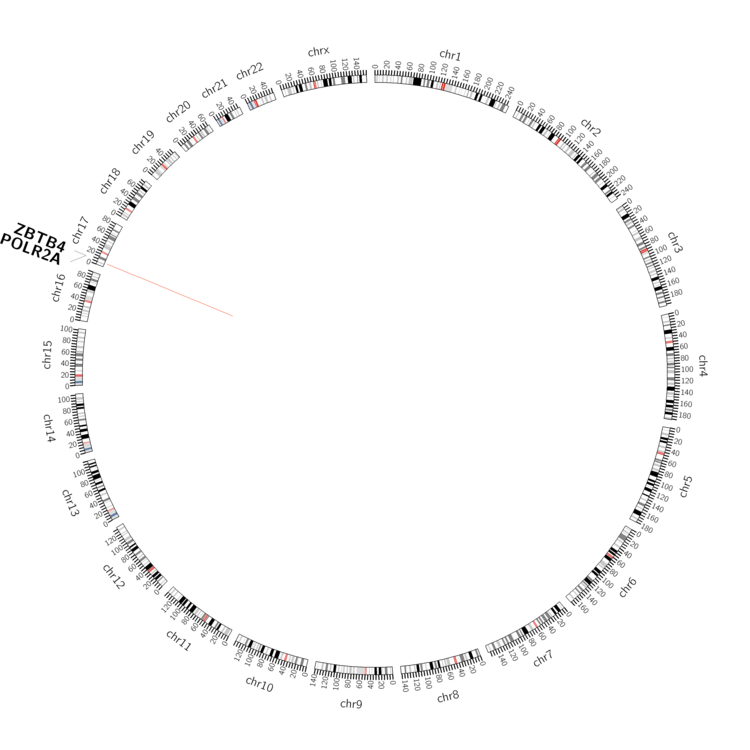 |
| - For Inter-chromosomal Variations |
| There's no inter-chromosomal structural variation. |
| - For Intra-chromosomal Variations |
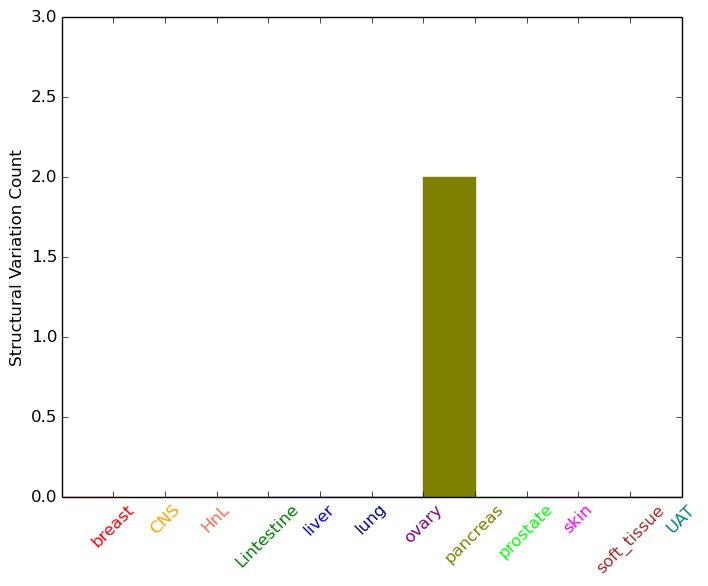
| * Intra-chromosomal variantions includes 'intrachromosomal amplicon to amplicon', 'intrachromosomal amplicon to non-amplified dna', 'intrachromosomal deletion', 'intrachromosomal fold-back inversion', 'intrachromosomal inversion', 'intrachromosomal tandem duplication', 'Intrachromosomal unknown type', 'intrachromosomal with inverted orientation', 'intrachromosomal with non-inverted orientation'. |
 |
| Sample | Symbol_a | Chr_a | Start_a | End_a | Symbol_b | Chr_b | Start_b | End_b |
| pancreas | POLR2A | chr17 | 7412656 | 7412676 | ZBTB4 | chr17 | 7383899 | 7383919 |
| cf) Tissue number; Tissue name (1;Breast, 2;Central_nervous_system, 3;Haematopoietic_and_lymphoid_tissue, 4;Large_intestine, 5;Liver, 6;Lung, 7;Ovary, 8;Pancreas, 9;Prostate, 10;Skin, 11;Soft_tissue, 12;Upper_aerodigestive_tract) |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows POLR2A related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| BE819347 | NUP188 | 3 | 151 | 9 | 131768587 | 131768868 | POLR2A | 138 | 287 | 17 | 7407022 | 7411630 | |
| BF805544 | CEP250 | 1 | 236 | 20 | 34092642 | 34095612 | POLR2A | 230 | 538 | 17 | 7416611 | 7416919 | |
| DA107051 | GPM6A | 1 | 248 | 4 | 176622745 | 176733415 | POLR2A | 249 | 581 | 17 | 7404138 | 7404883 | |
| BI061851 | IKZF1 | 1 | 162 | 7 | 50424791 | 50424953 | POLR2A | 151 | 344 | 17 | 7404378 | 7404893 | |
| BF514571 | POLR2A | 18 | 79 | 17 | 7400767 | 7400828 | POLR2A | 74 | 285 | 17 | 7401012 | 7401430 | |
| BF512093 | POLR2A | 18 | 79 | 17 | 7400767 | 7400828 | POLR2A | 74 | 285 | 17 | 7401012 | 7401430 | |
| BI059477 | IKZF1 | 1 | 162 | 7 | 50424791 | 50424953 | POLR2A | 151 | 344 | 17 | 7404378 | 7404893 | |
| BE833427 | USP12 | 5 | 28 | 13 | 27666061 | 27666087 | POLR2A | 18 | 258 | 17 | 7416128 | 7416459 | |
| BE819348 | NUP188 | 13 | 176 | 9 | 131768570 | 131768868 | POLR2A | 163 | 312 | 17 | 7407022 | 7411630 | |
| BI022194 | AP2B1 | 1 | 211 | 17 | 33951591 | 33953833 | POLR2A | 203 | 437 | 17 | 7403972 | 7404297 | |
| BE159844 | POLR2A | 39 | 60 | 17 | 7409231 | 7409252 | PVRL2 | 45 | 382 | 19 | 45376694 | 45377031 | |
| Top |
| Mutation type/ Tissue ID | brca | cns | cerv | endome | haematopo | kidn | Lintest | liver | lung | ns | ovary | pancre | prost | skin | stoma | thyro | urina | |||
| Total # sample | 1 | |||||||||||||||||||
| GAIN (# sample) | 1 | |||||||||||||||||||
| LOSS (# sample) |
| cf) Tissue ID; Tissue type (1; Breast, 2; Central_nervous_system, 3; Cervix, 4; Endometrium, 5; Haematopoietic_and_lymphoid_tissue, 6; Kidney, 7; Large_intestine, 8; Liver, 9; Lung, 10; NS, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary_tract) |
| Top |
|
 |
| Top |
| Stat. for Non-Synonymous SNVs (# total SNVs=108) | (# total SNVs=48) |
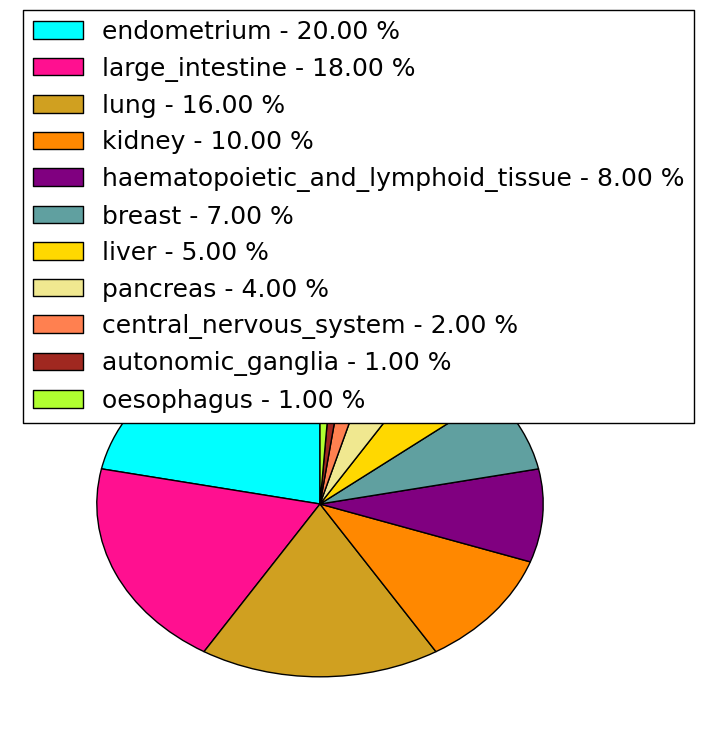 | 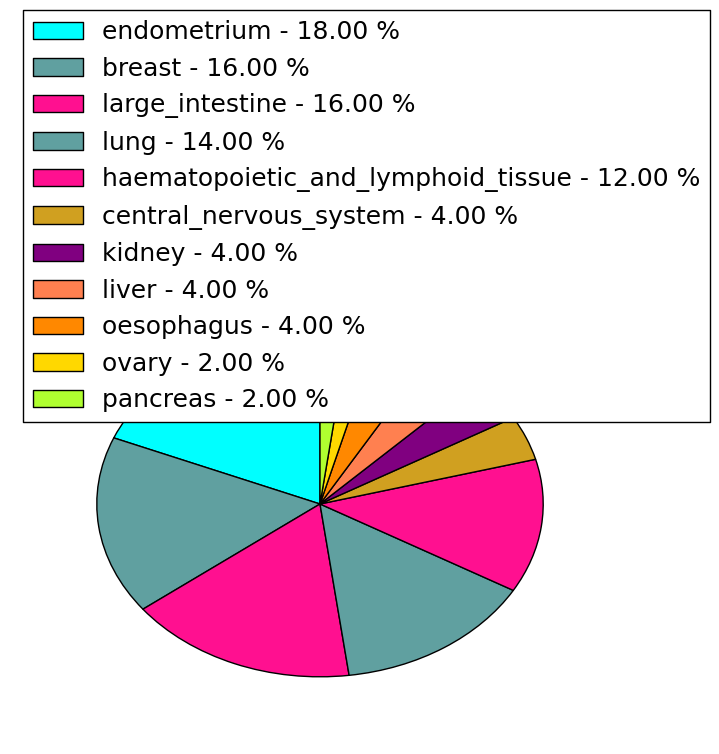 |
(# total SNVs=6) | (# total SNVs=1) |
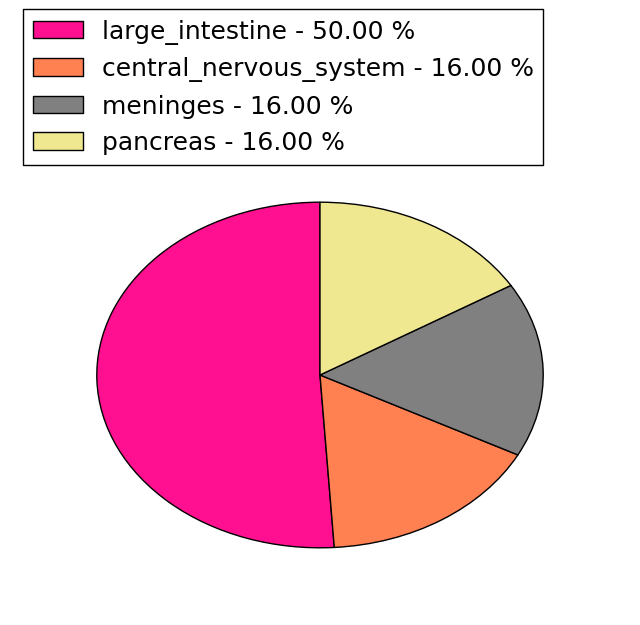 | 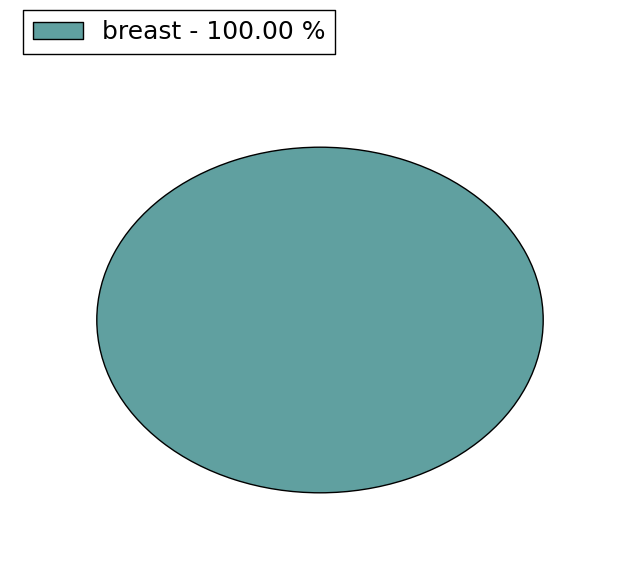 |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr17:7416599-7416599 | p.S1672S | 4 |
| chr17:7412911-7412911 | p.R1258H | 3 |
| chr17:7405316-7405316 | p.G816V | 3 |
| chr17:7399866-7399866 | p.G157G | 3 |
| chr17:7401440-7401440 | p.A416T | 3 |
| chr17:7416909-7416909 | p.Y1776H | 2 |
| chr17:7405965-7405965 | p.V901F | 2 |
| chr17:7405418-7405418 | p.T850K | 2 |
| chr17:7417385-7417385 | p.S1934S | 2 |
| chr17:7400127-7400127 | p.S194S | 2 |
| Top |
|
 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 5 | 1 | 17 | 2 | 11 | 7 | 2 | 14 | 4 | 1 | 1 | 18 | 18 | 1 | 16 | |||||
| # mutation | 5 | 1 | 17 | 2 | 11 | 7 | 2 | 15 | 4 | 1 | 1 | 22 | 22 | 1 | 28 | |||||
| nonsynonymous SNV | 4 | 11 | 1 | 9 | 6 | 2 | 10 | 3 | 1 | 13 | 17 | 1 | 20 | |||||||
| synonymous SNV | 1 | 1 | 6 | 1 | 2 | 1 | 5 | 1 | 1 | 9 | 5 | 8 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr17:7401440 | p.G150D | 3 |
| chr17:7399844 | p.A416T | 3 |
| chr17:7404318 | p.R1603H | 2 |
| chr17:7416821 | p.C451C | 2 |
| chr17:7402375 | p.T647T | 2 |
| chr17:7416391 | p.R475H | 2 |
| chr17:7402446 | p.L1956L | 2 |
| chr17:7417451 | p.P1746P | 2 |
| chr17:7411775 | p.S1829S | 1 |
| chr17:7416688 | p.R604H | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
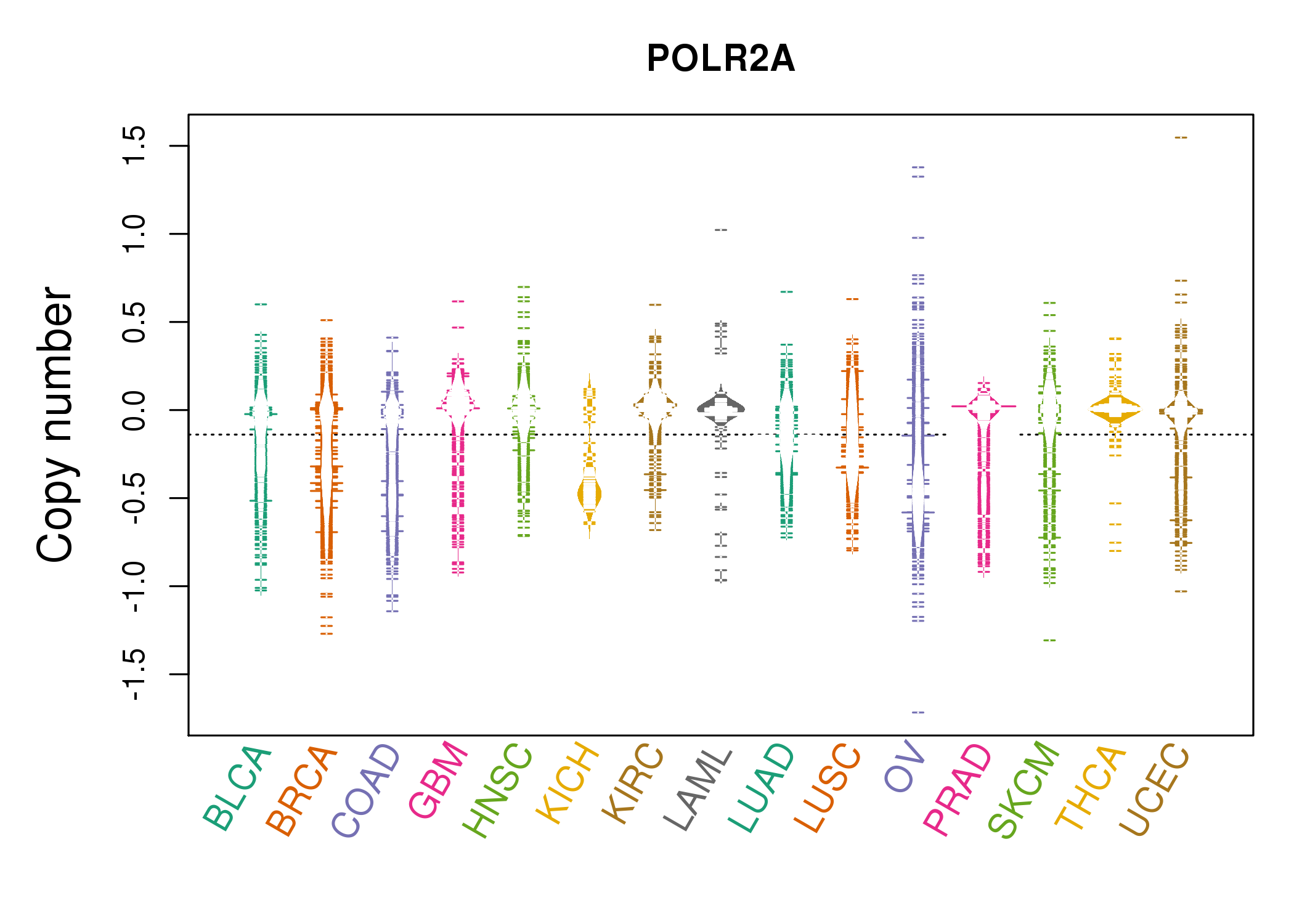 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
| Gene Expression for POLR2A |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
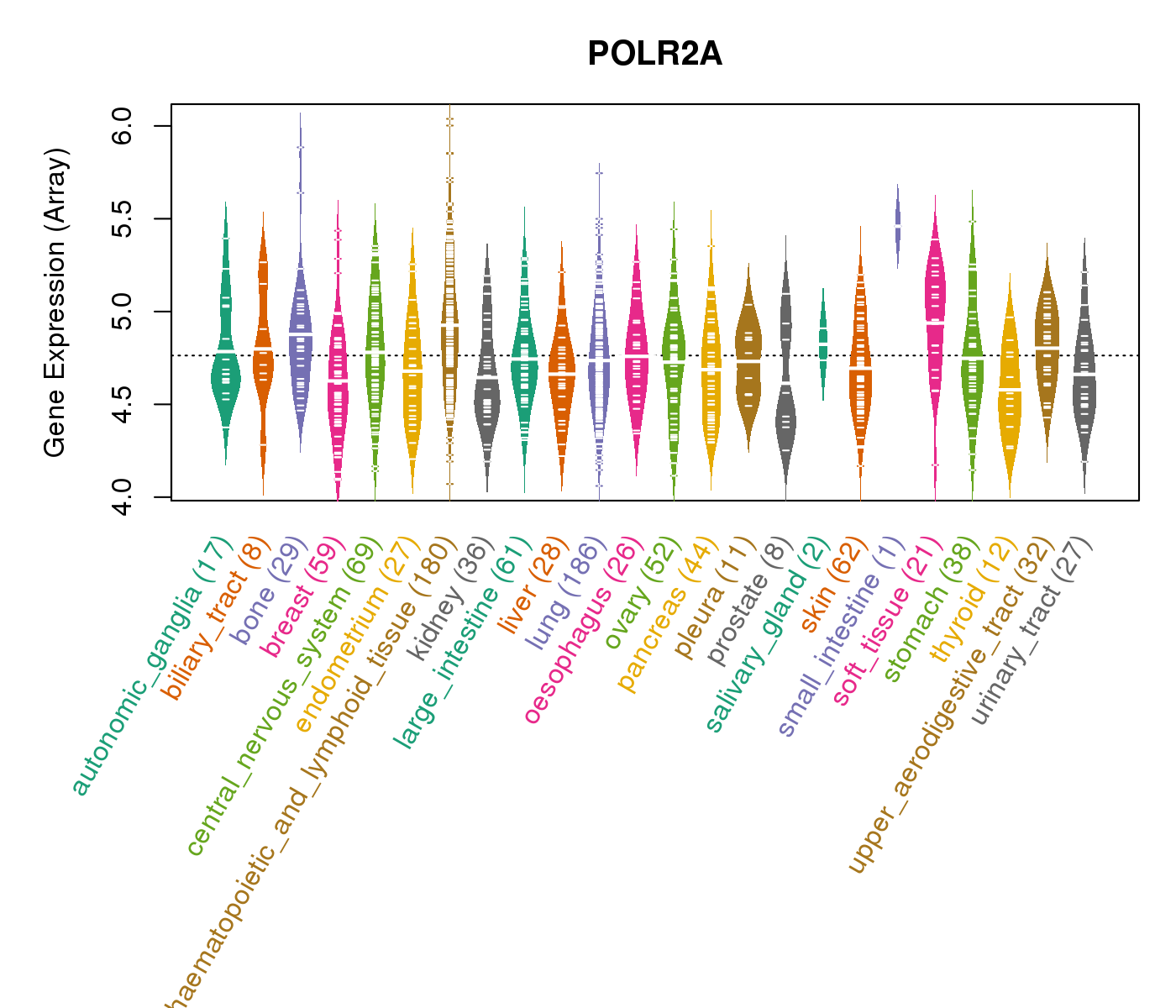 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
 |
| Top |
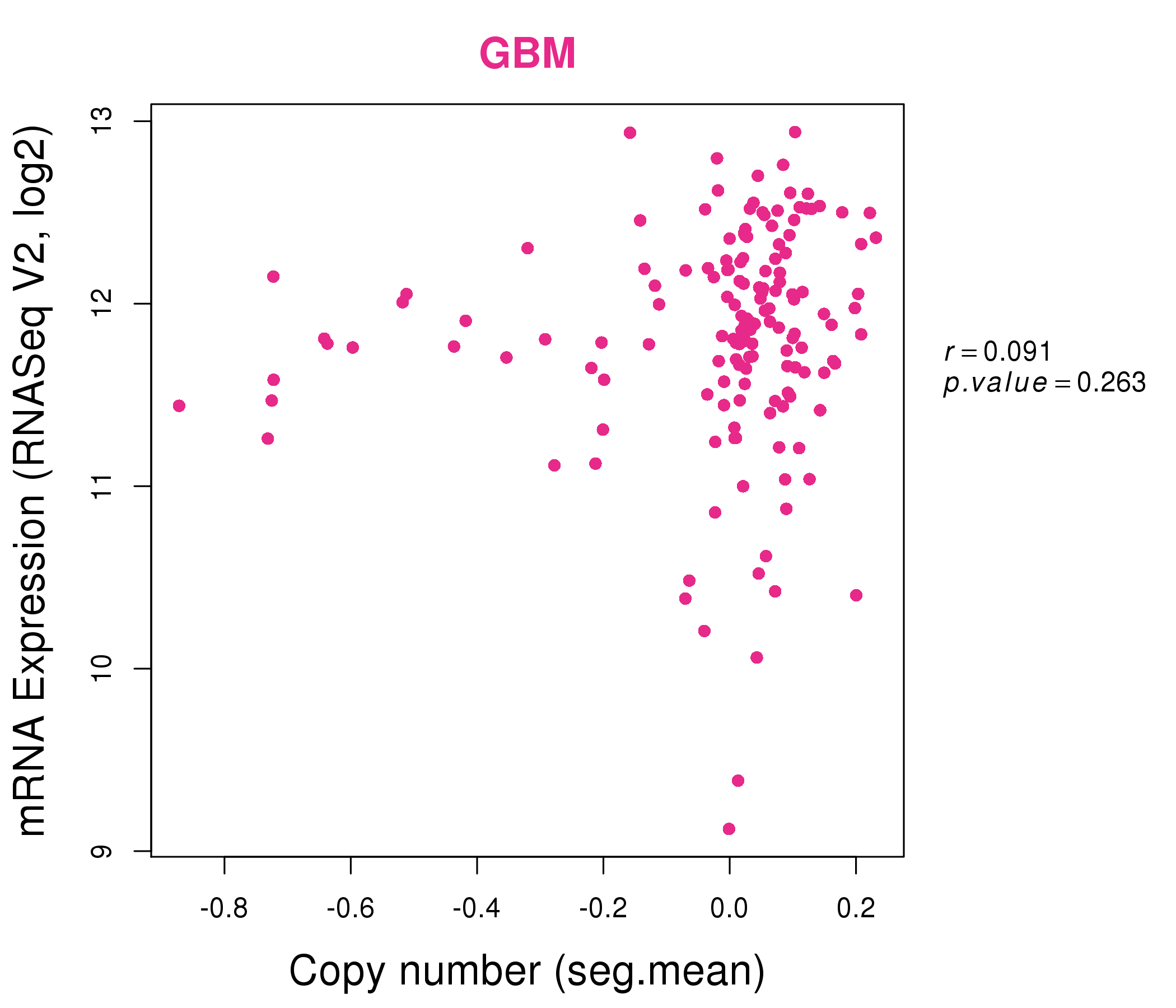
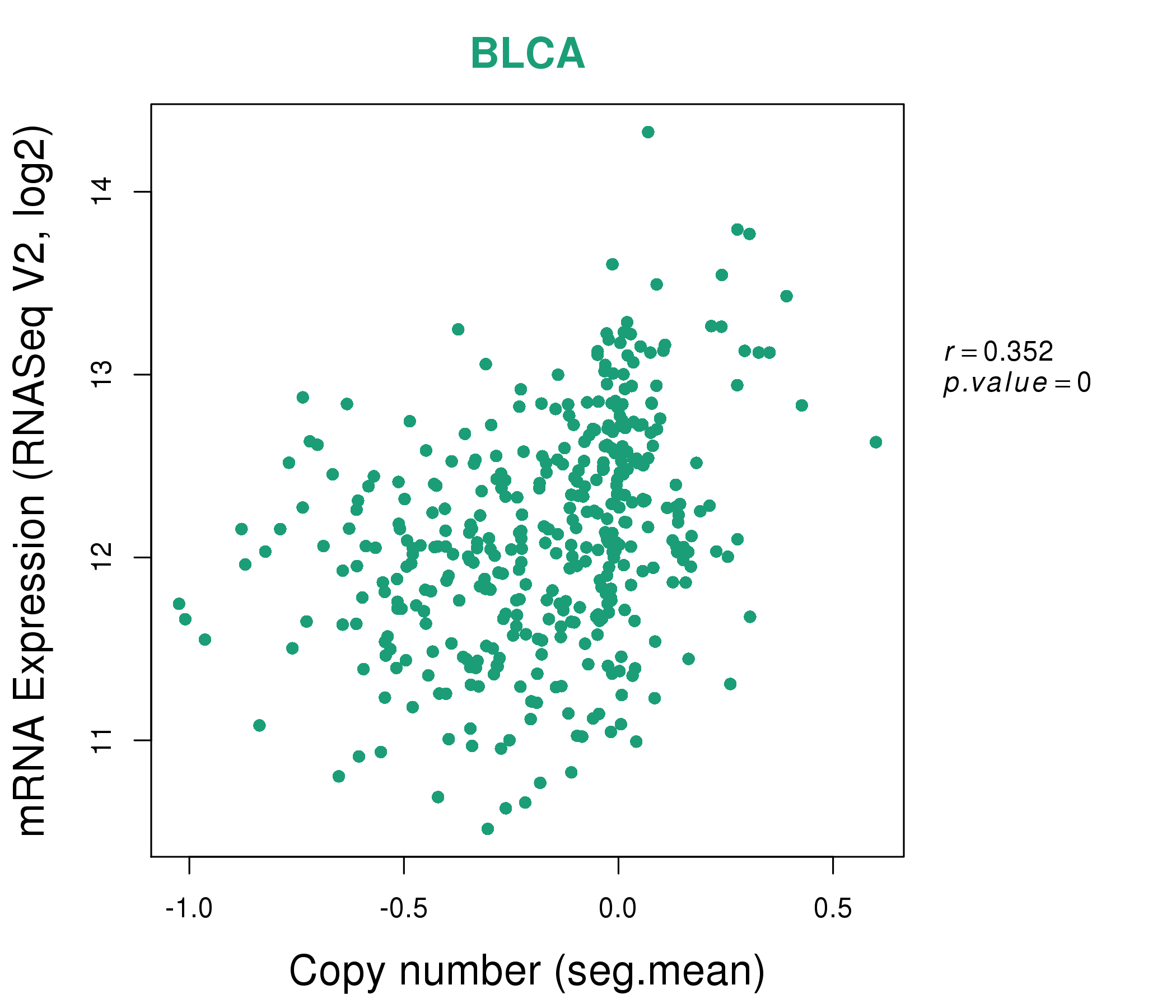
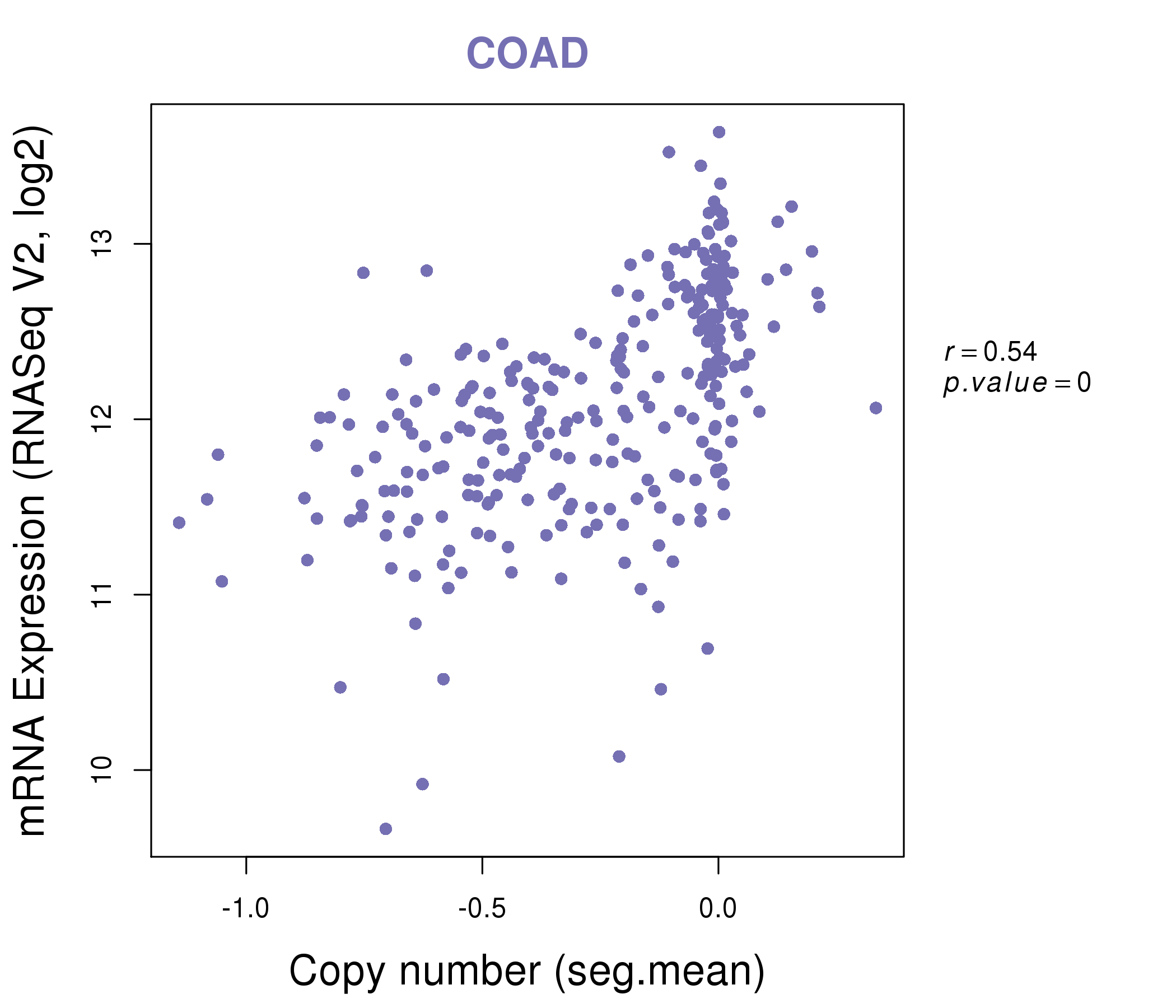
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
 |
|
 |
|
| Top |
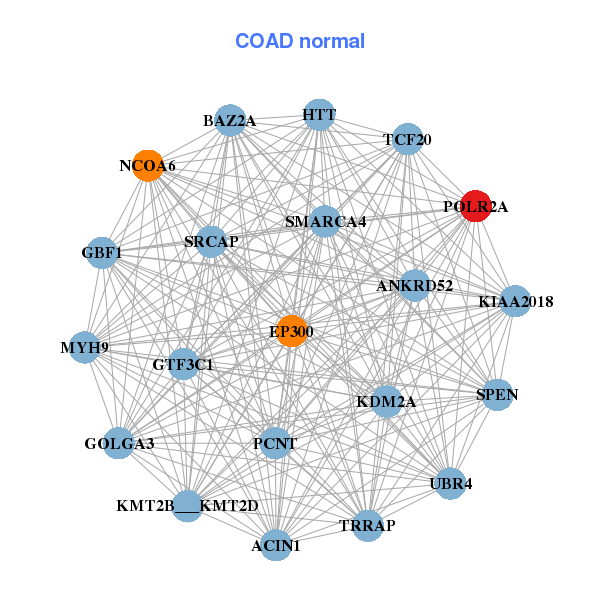
| Gene-Gene Network Information |
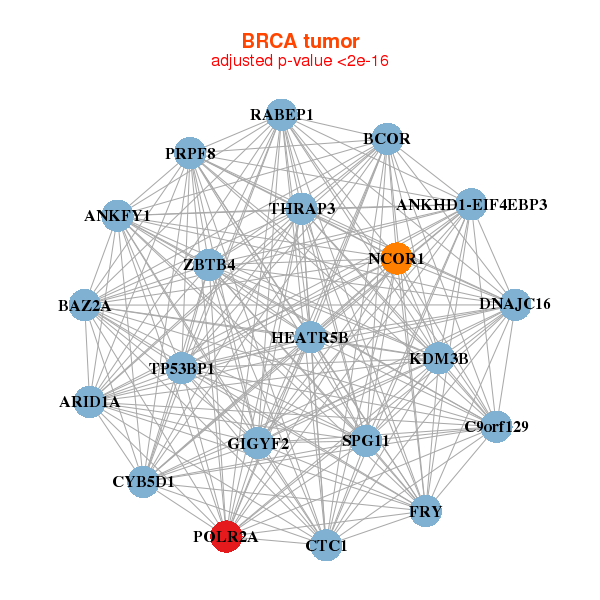
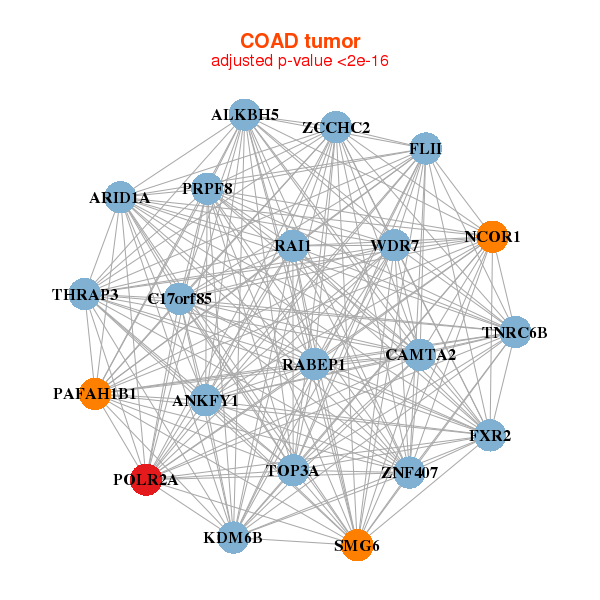
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
 |
| ||||
| ANKFY1,ANKHD1-EIF4EBP3,ARID1A,BAZ2A,BCOR,CTC1,C9orf129, CYB5D1,DNAJC16,FRY,GIGYF2,HEATR5B,KDM3B,NCOR1, POLR2A,PRPF8,RABEP1,SPG11,THRAP3,TP53BP1,ZBTB4 | ARID1A,BRPF3,C2CD3,COL4A5,EP400,KIAA0556,KIAA1217, KIAA1549,MAML1,MAP3K9,MDC1,POLR2A,SIPA1L1,SIPA1L3, SRCAP,TRRAP,TTLL5,YLPM1,ZNF135,ZNF142,ZSWIM5 | ||||
 |
| ||||
| ALKBH5,ANKFY1,ARID1A,C17orf85,CAMTA2,FLII,FXR2, KDM6B,NCOR1,PAFAH1B1,POLR2A,PRPF8,RABEP1,RAI1, SMG6,THRAP3,TNRC6B,TOP3A,WDR7,ZCCHC2,ZNF407 | ACIN1,ANKRD52,BAZ2A,EP300,GBF1,GOLGA3,GTF3C1, HTT,KDM2A,KIAA2018,KMT2B___KMT2D,MYH9,NCOA6,PCNT, POLR2A,SMARCA4,SPEN,SRCAP,TCF20,TRRAP,UBR4 |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for POLR2A |
| DB Category | DB Name | DB's ID and Url link |
| Chemistry | BindingDB | P24928; -. |
| Chemistry | ChEMBL | CHEMBL1641353; -. |
| Organism-specific databases | PharmGKB | PA33507; -. |
| Organism-specific databases | CTD | 5430; -. |
| * Gene Centered Interaction Network. |
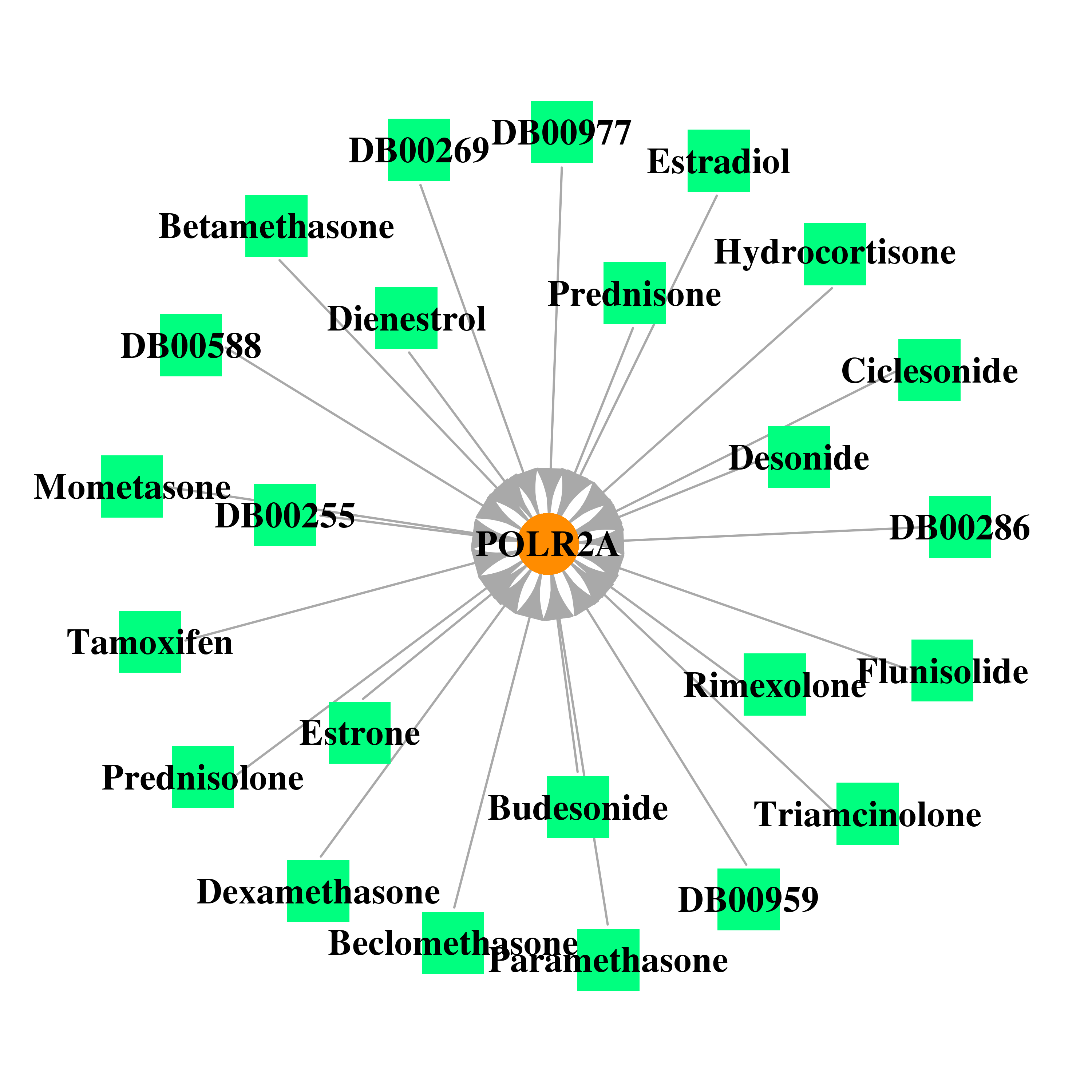 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB00269 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Chlorotrianisene | 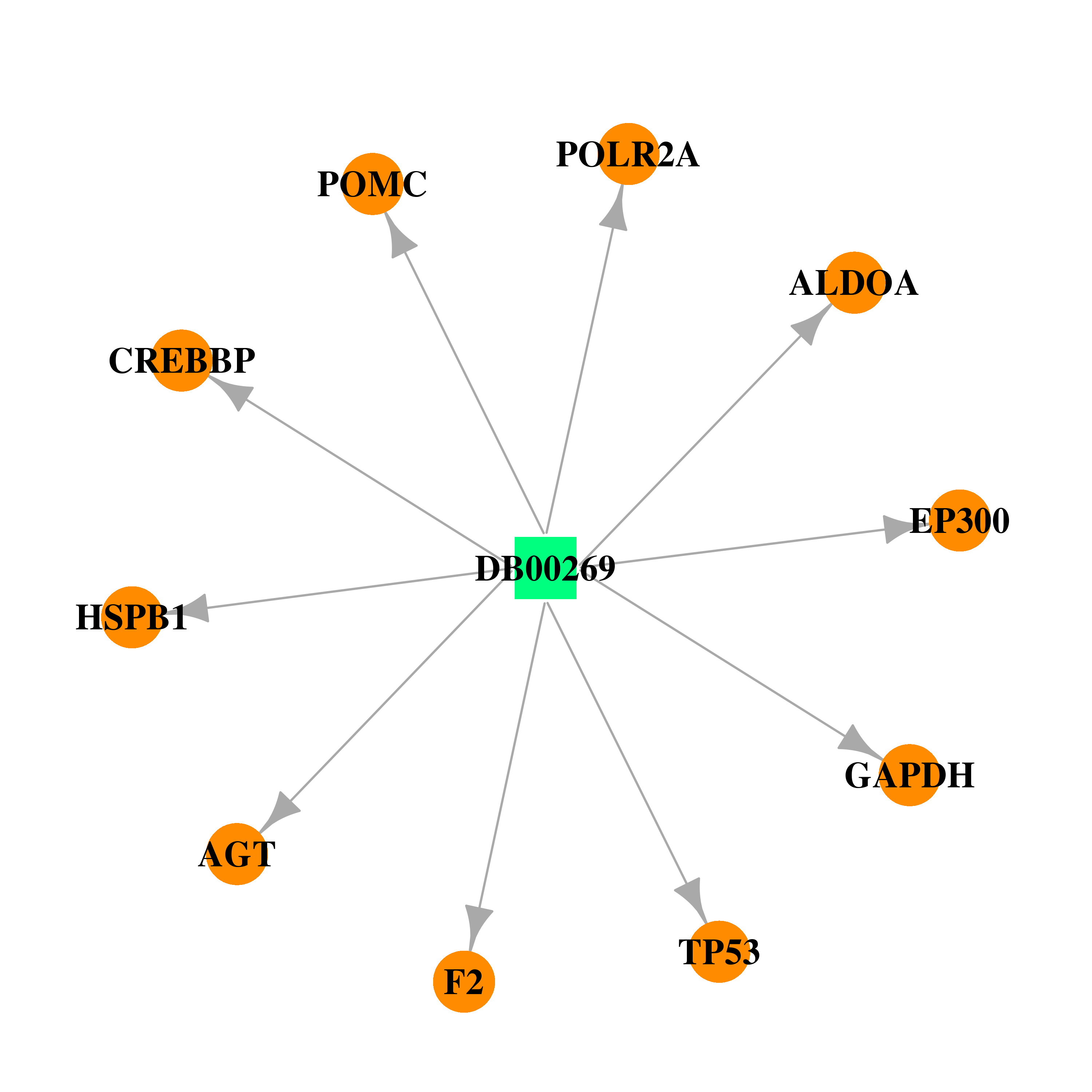 |  |
| DB00286 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Conjugated Estrogens | 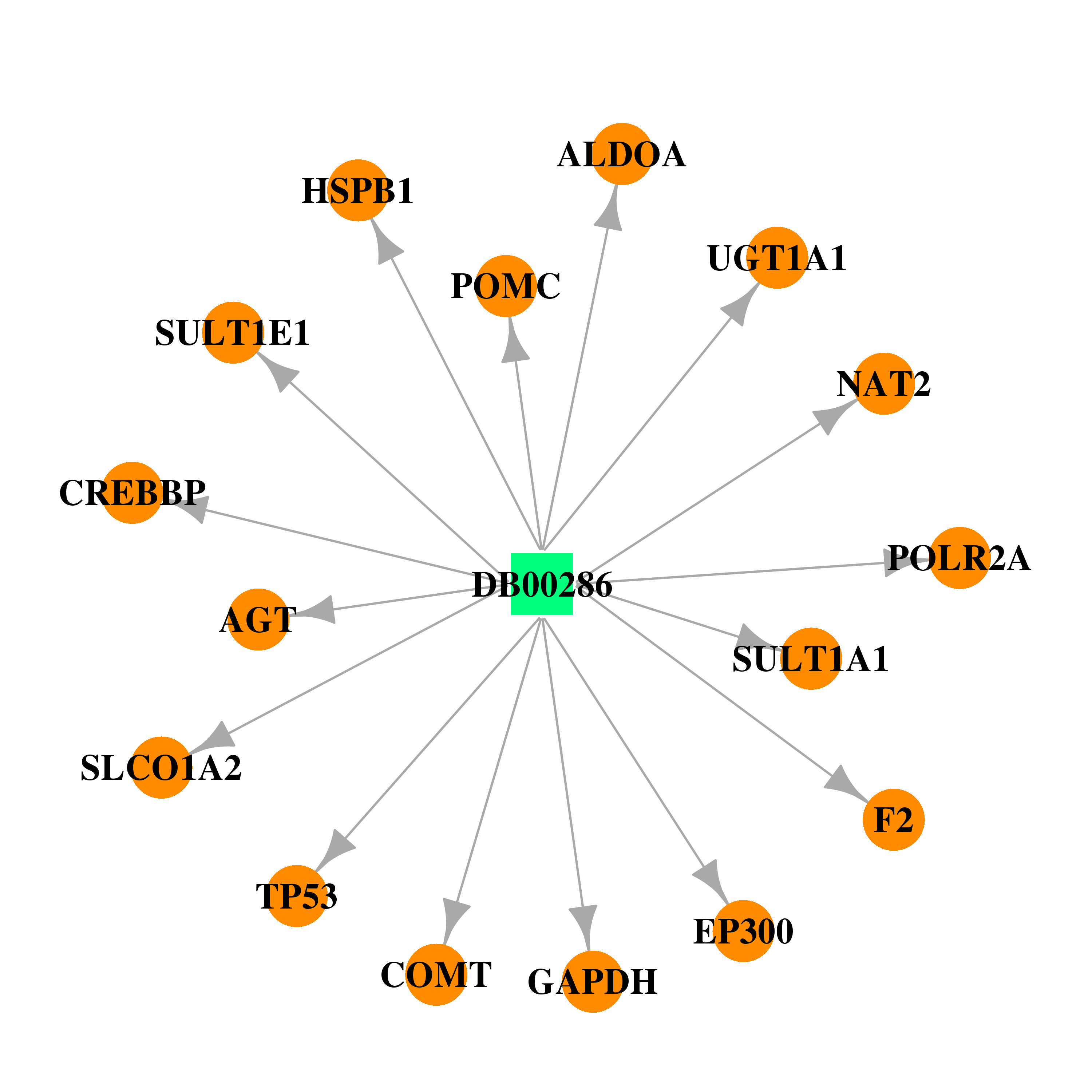 |  |
| DB00255 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Diethylstilbestrol |  |  |
| DB00783 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved; investigational | Estradiol | 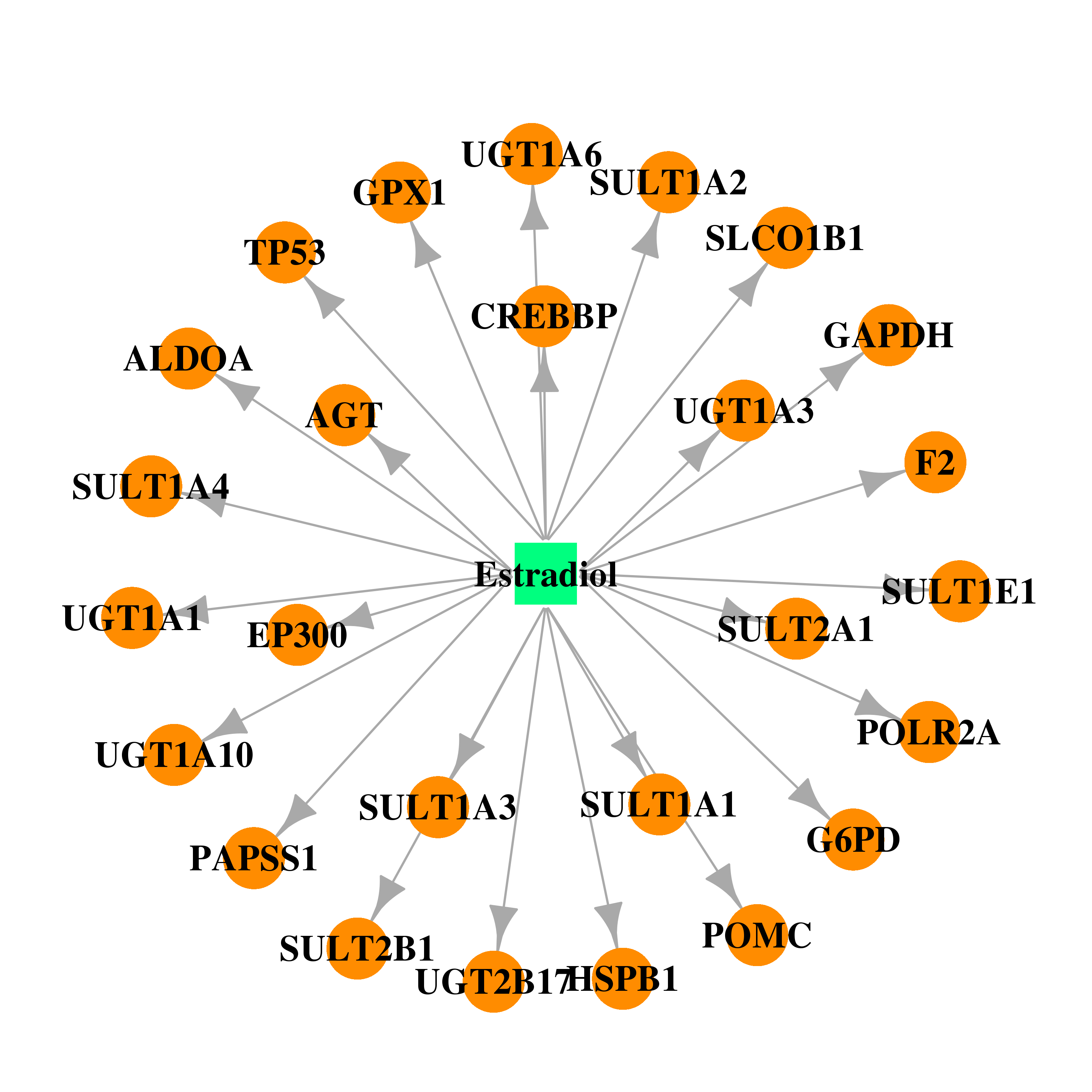 |  |
| DB00655 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Estrone |  |  |
| DB00977 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Ethinyl Estradiol |  |  |
| DB00890 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Dienestrol |  | 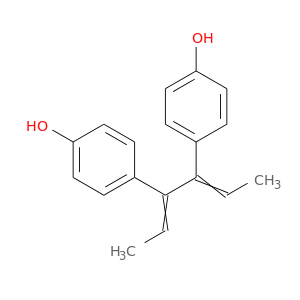 |
| DB00675 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Tamoxifen |  |  |
| DB00394 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Beclomethasone | 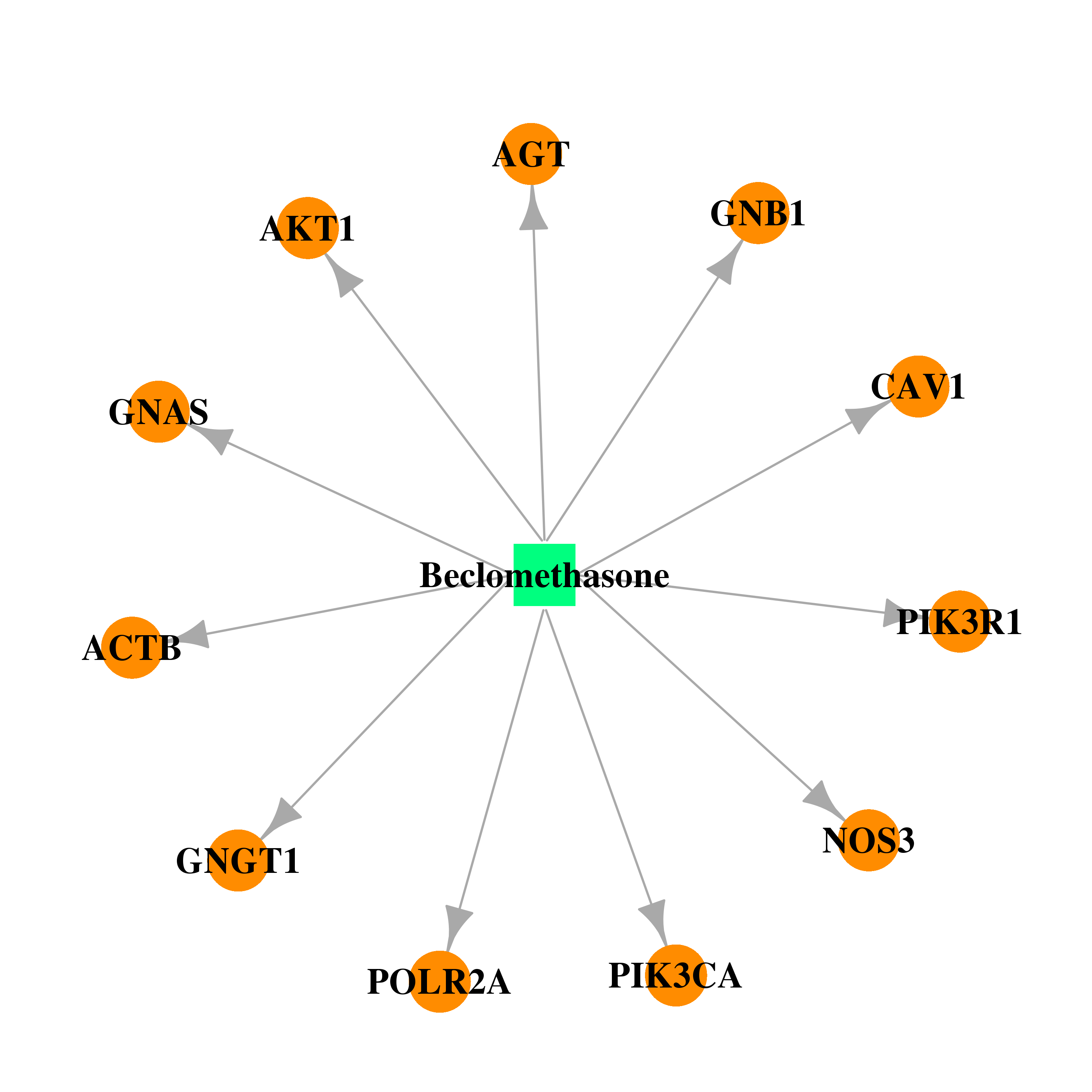 | 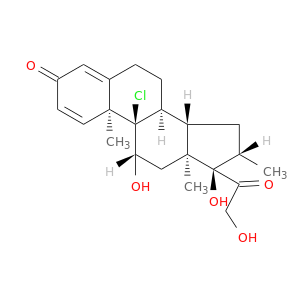 |
| DB00443 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Betamethasone |  | 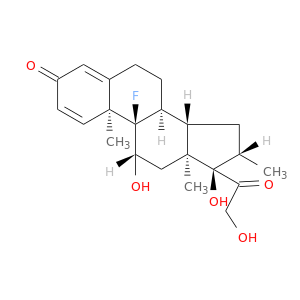 |
| DB01222 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved; investigational | Budesonide |  |  |
| DB01410 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved; investigational | Ciclesonide | 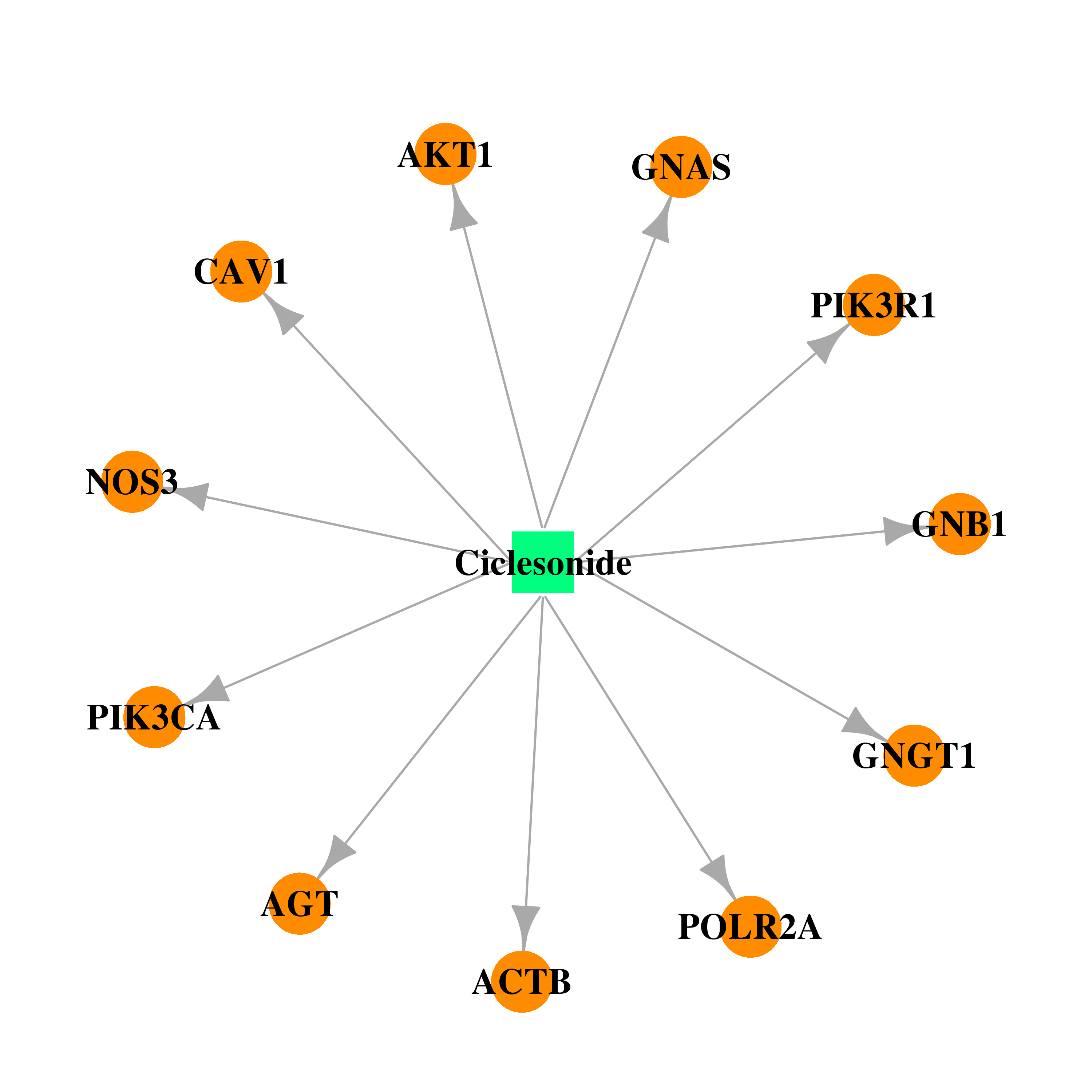 |  |
| DB01260 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved; investigational | Desonide |  |  |
| DB01234 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved; investigational | Dexamethasone | 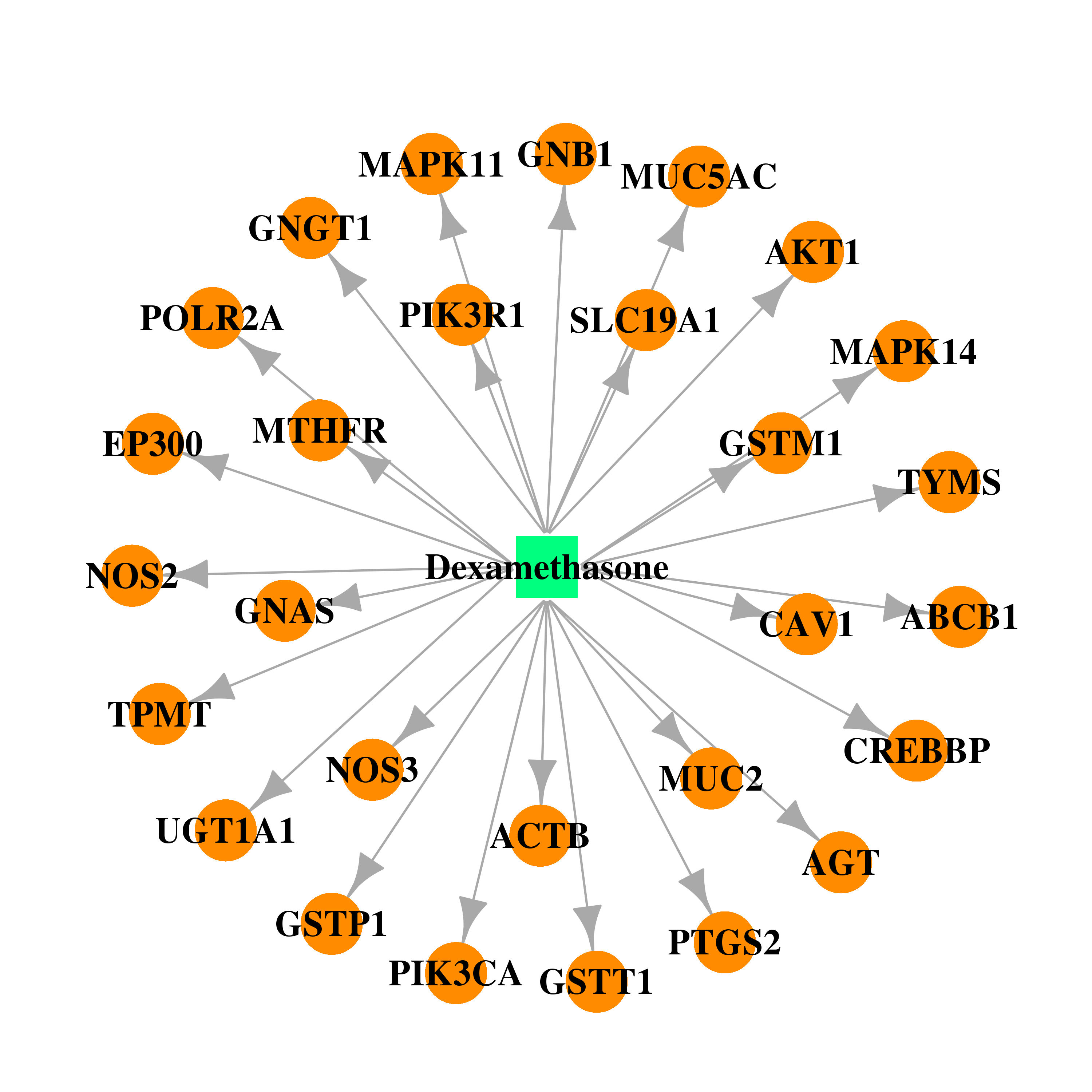 | 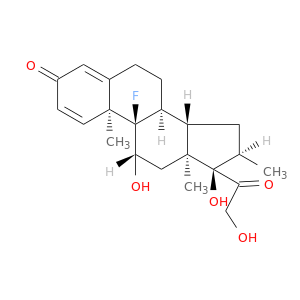 |
| DB00180 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved; investigational | Flunisolide | 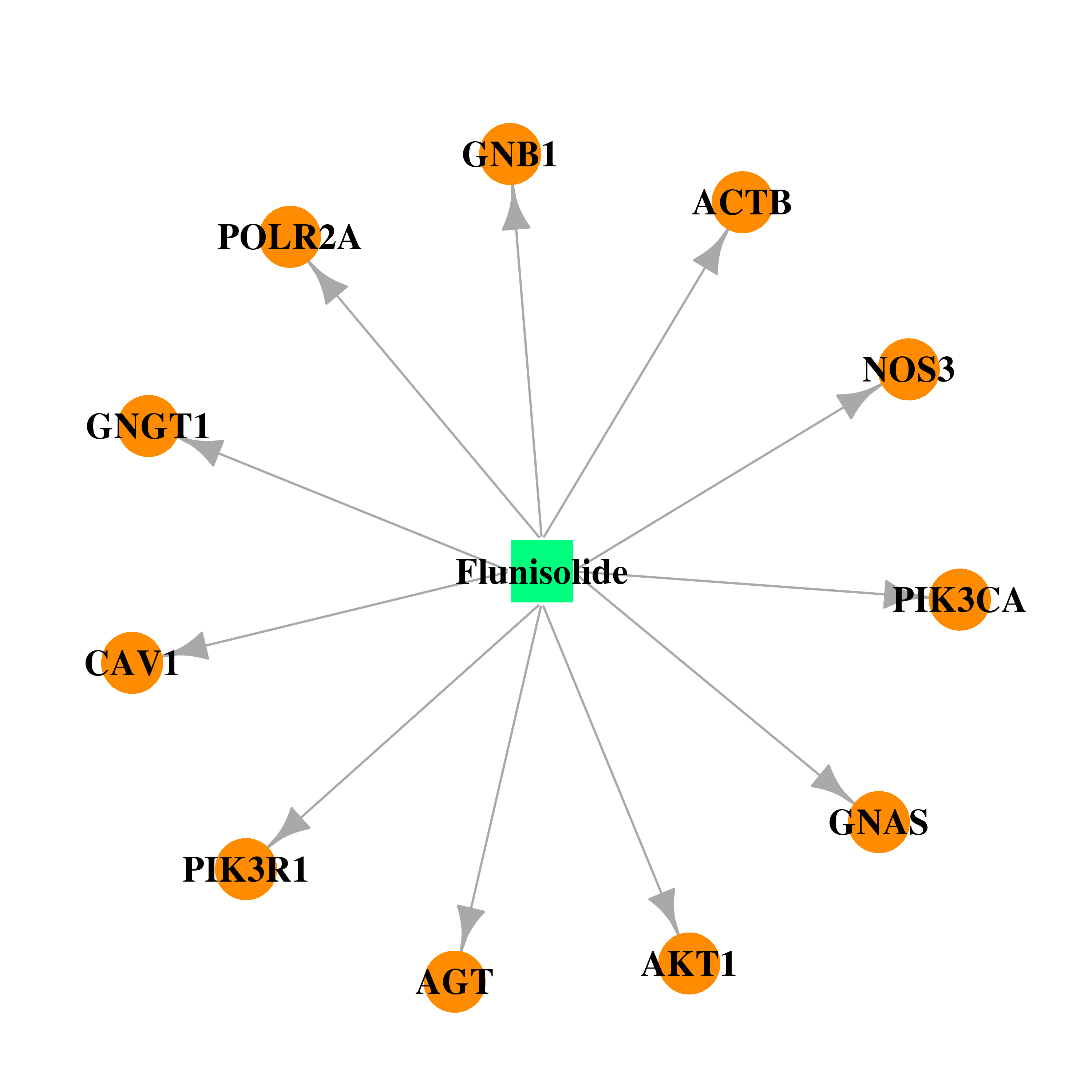 | 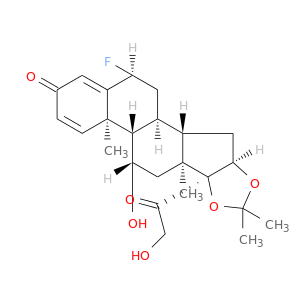 |
| DB00588 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved; investigational | Fluticasone Propionate | 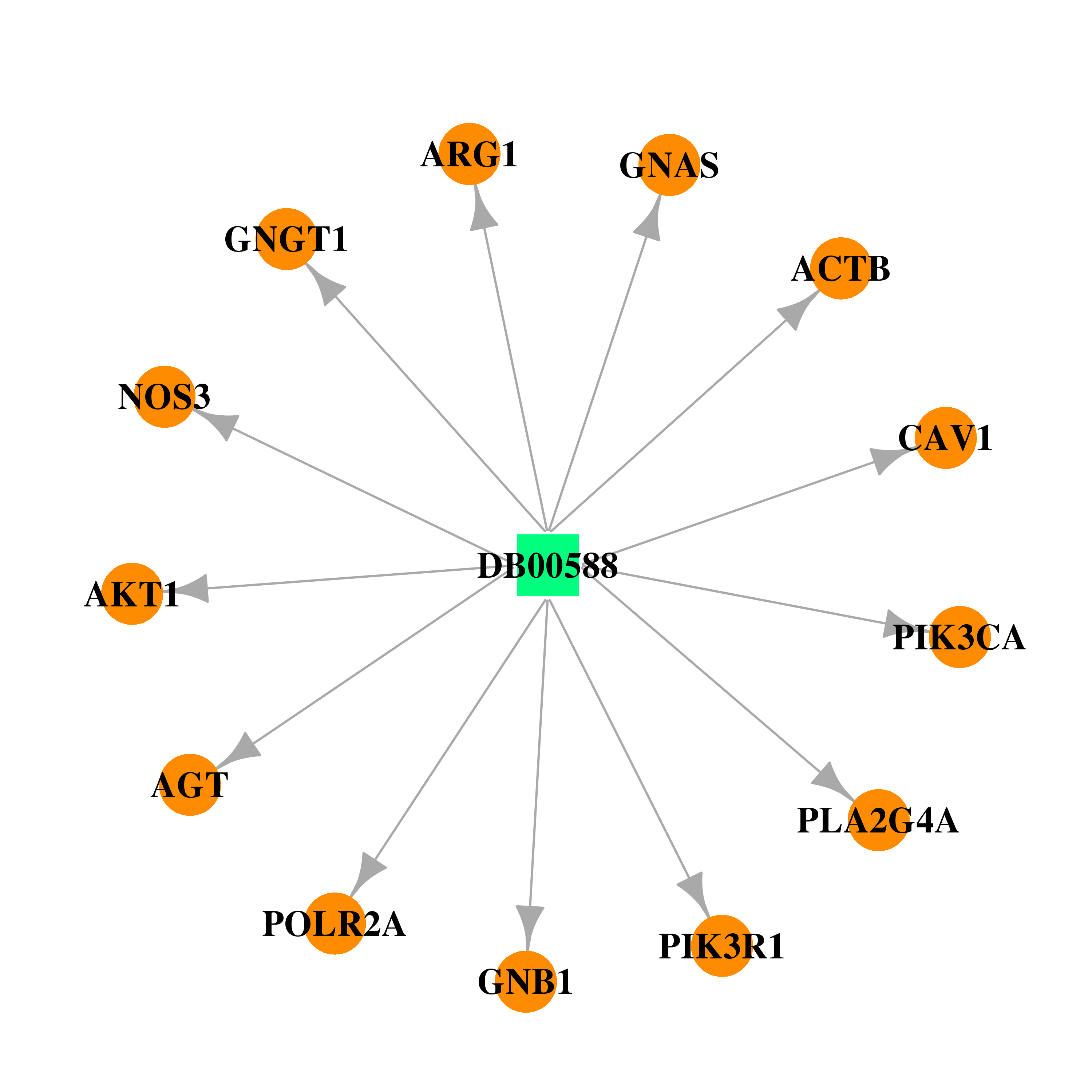 |  |
| DB00741 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Hydrocortisone |  |  |
| DB00959 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Methylprednisolone |  |  |
| DB00764 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Mometasone | 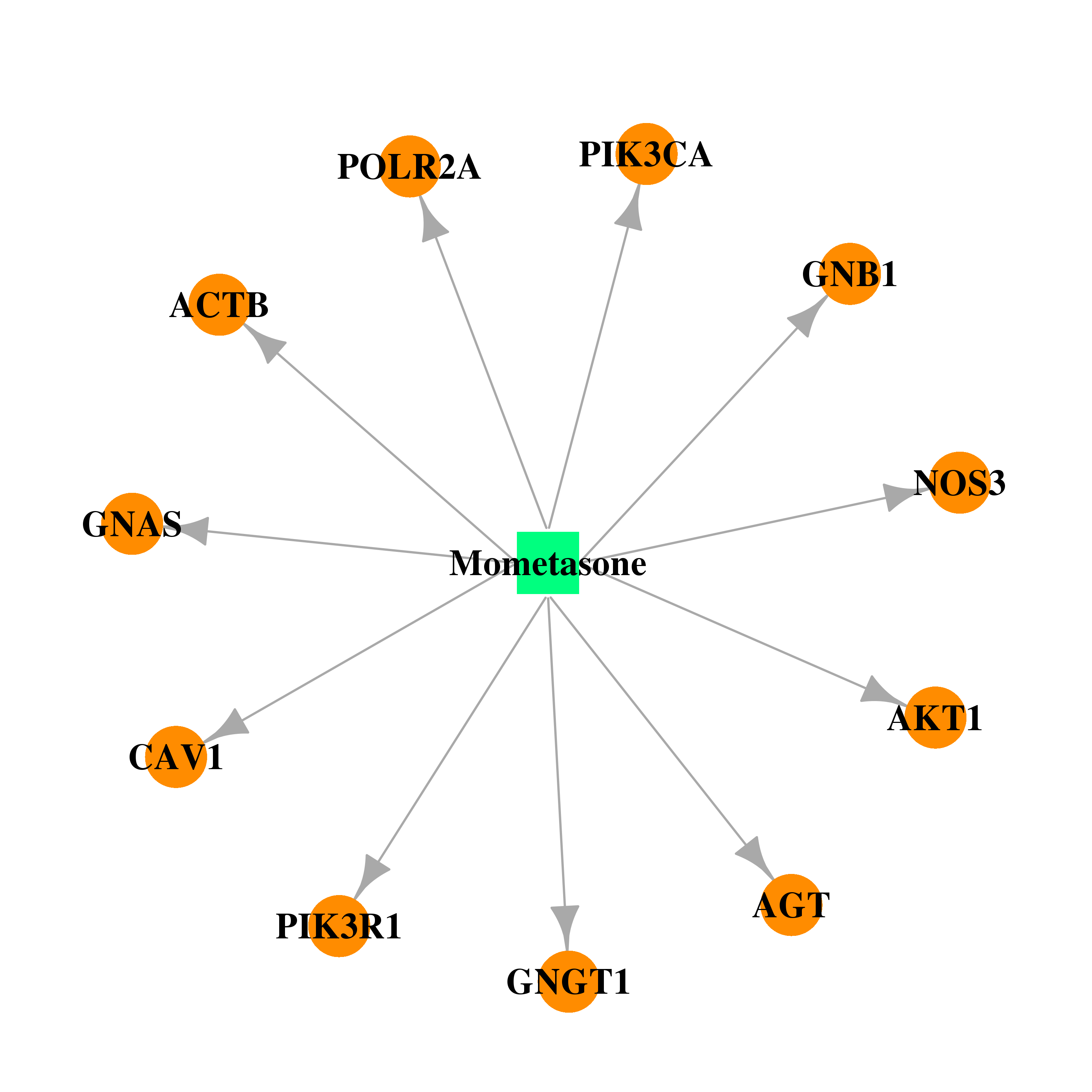 |  |
| DB01384 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Paramethasone | 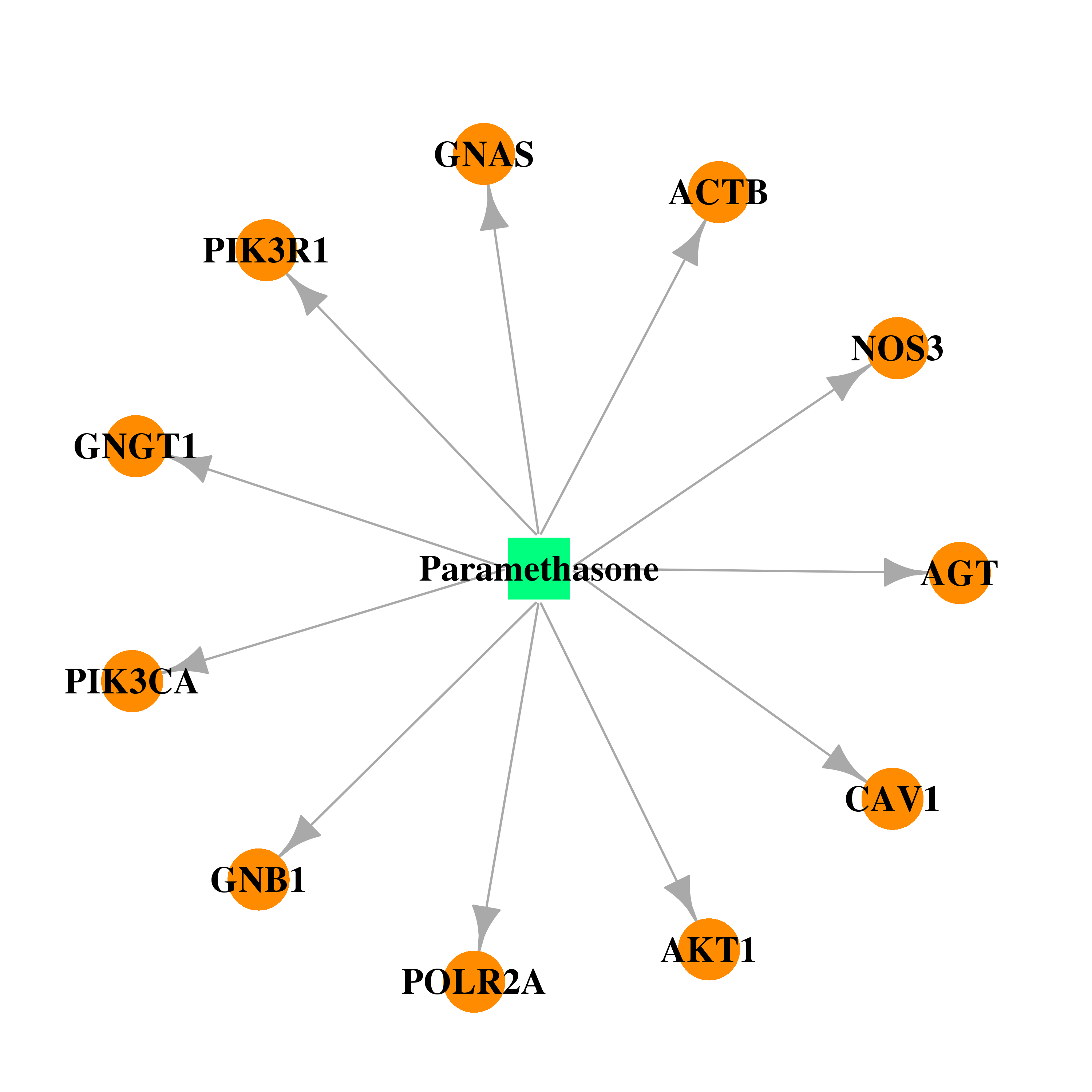 |  |
| DB00860 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Prednisolone | 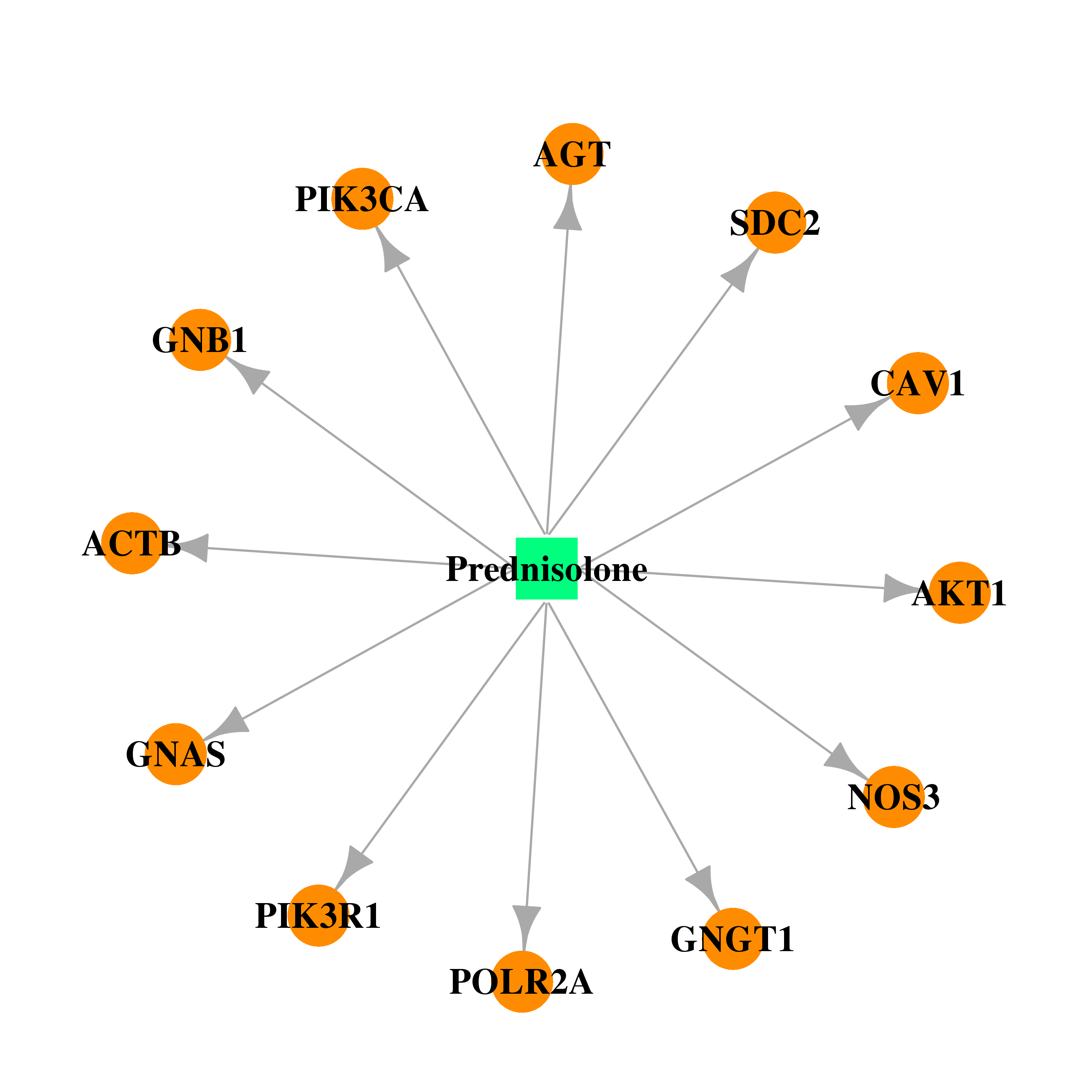 |  |
| DB00635 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Prednisone | 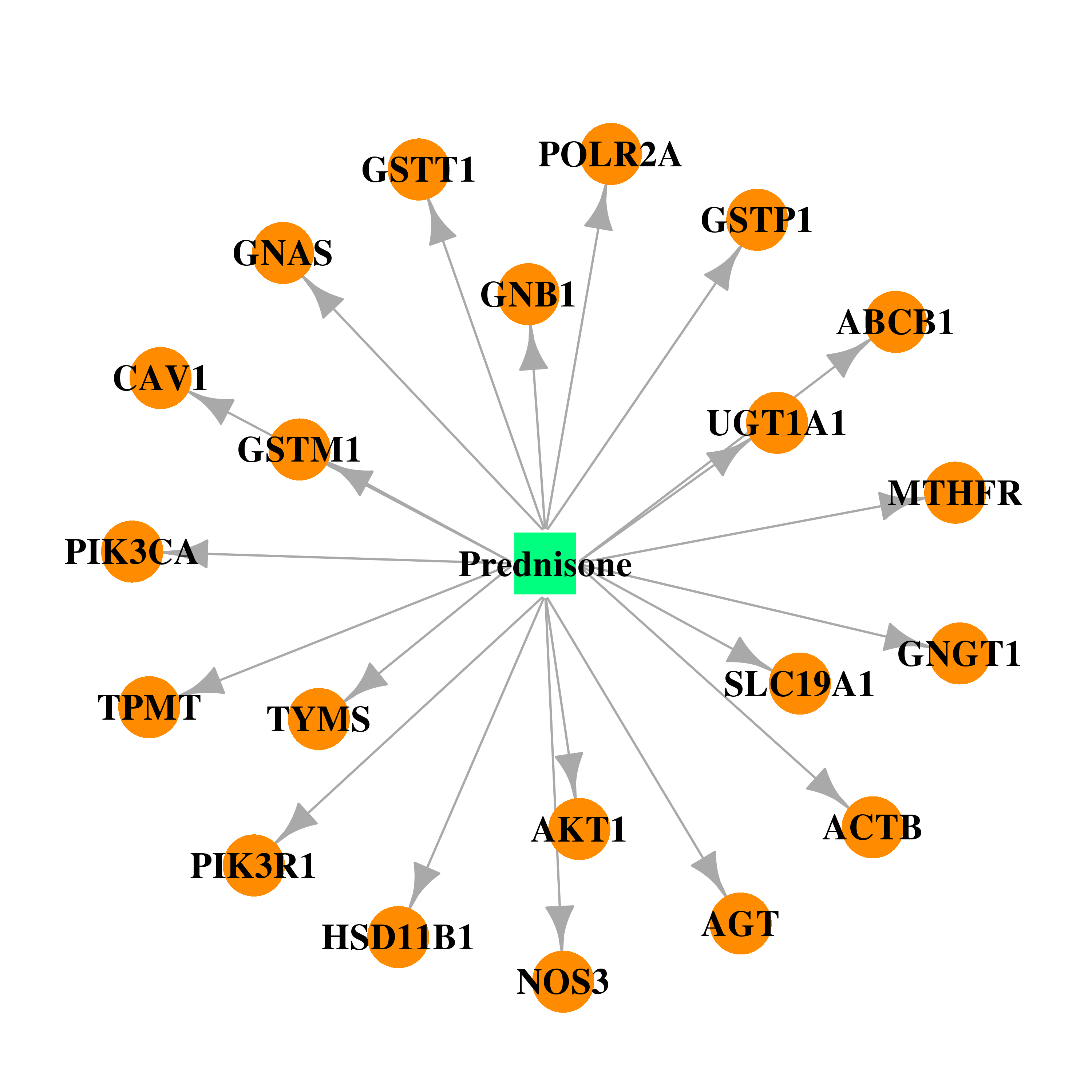 |  |
| DB00896 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Rimexolone |  |  |
| DB00620 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | approved | Triamcinolone |  |  |
| Top |
| Cross referenced IDs for POLR2A |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |