| Ligand ID | Ligand short name | Ligand long name | PDB ID | PDB name | mutLBS |
NMN | | NICOTINAMIDE MONONUCLEOTIDE | 2gvg | A | D354 | NMN | | NICOTINAMIDE MONONUCLEOTIDE | 2gvg | C | D354 | NMN | | NICOTINAMIDE MONONUCLEOTIDE | 2gvg | D | D354 | NMN | | NICOTINAMIDE MONONUCLEOTIDE | 2gvg | E | D354 | PRP | | 5-O-PHOSPHONO-ALPHA-D-RIBOFURANOSYL DIPHOSPHATE | 3dkj | B | D354 | DGB | | N-[4-(1-BENZOYLPIPERIDIN-4-YL)BUTYL]-3-PYRIDIN-3-YLPROPANAMIDE | 4o1b | A | G185 H191 S275 | DGB | | N-[4-(1-BENZOYLPIPERIDIN-4-YL)BUTYL]-3-PYRIDIN-3-YLPROPANAMIDE | 4o1b | B | G185 H191 S275 | 20J | | 7-CHLORO-3-METHYL-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE | 4lv9 | A | H191 | 20J | | 7-CHLORO-3-METHYL-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE | 4lv9 | B | H191 | 1EB | | N-(4-NITROPHENYL)CYCLOPROPANECARBOXAMIDE | 4lvd | B | H191 | 1EB | | N-(4-NITROPHENYL)CYCLOPROPANECARBOXAMIDE | 4lvd | A | H191 | DGB | | N-[4-(1-BENZOYLPIPERIDIN-4-YL)BUTYL]-3-PYRIDIN-3-YLPROPANAMIDE | 2gvj | A | H191 S275 | 1LS | | 1-[4-(PIPERIDIN-1-YLSULFONYL)PHENYL]-3-(PYRIDIN-3- YLMETHYL)THIOUREA | 4jr5 | A | H191 S275 | 1LS | | 1-[4-(PIPERIDIN-1-YLSULFONYL)PHENYL]-3-(PYRIDIN-3- YLMETHYL)THIOUREA | 4jr5 | B | H191 S275 | 1QR | | N-[4-(PIPERIDIN-1-YLSULFONYL)BENZYL]-1H-PYRROLO[3,2- C]PYRIDINE-2-CARBOXAMIDE | 4kfn | A | H191 S275 | 1QR | | N-[4-(PIPERIDIN-1-YLSULFONYL)BENZYL]-1H-PYRROLO[3,2- C]PYRIDINE-2-CARBOXAMIDE | 4kfn | B | H191 S275 | 1QS | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}IMIDAZO[1,2- A]PYRIDINE-6-CARBOXAMIDE | 4kfo | A | H191 S275 | 1QS | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}IMIDAZO[1,2- A]PYRIDINE-6-CARBOXAMIDE | 4kfo | B | H191 S275 | 1LJ | | 1-[(6-AMINOPYRIDIN-3-YL)METHYL]-3-[4-(PHENYLSULFONYL) PHENYL]UREA | 4jnm | A | H191 S275 | 1LJ | | 1-[(6-AMINOPYRIDIN-3-YL)METHYL]-3-[4-(PHENYLSULFONYL) PHENYL]UREA | 4jnm | B | H191 S275 | 1R7 | | N-(4-{[1-(TETRAHYDRO-2H-PYRAN-4-YL)PIPERIDIN-4- YL]SULFONYL}BENZYL)-2H-PYRROLO[3,4-C]PYRIDINE-2- CARBOXAMIDE | 4kfp | A | H191 S275 | 1R7 | | N-(4-{[1-(TETRAHYDRO-2H-PYRAN-4-YL)PIPERIDIN-4- YL]SULFONYL}BENZYL)-2H-PYRROLO[3,4-C]PYRIDINE-2- CARBOXAMIDE | 4kfp | B | H191 S275 | 20M | | N-(4-{[4-(PYRROLIDIN-1-YL)PIPERIDIN-1- YL]SULFONYL}BENZYL)-2H-PYRIDO[4,3-E][1,2,4]THIADIAZIN- 3-AMINE 1,1-DIOXIDE | 4lva | A | H191 S275 | 20M | | N-(4-{[4-(PYRROLIDIN-1-YL)PIPERIDIN-1- YL]SULFONYL}BENZYL)-2H-PYRIDO[4,3-E][1,2,4]THIADIAZIN- 3-AMINE 1,1-DIOXIDE | 4lva | B | H191 S275 | 20N | | N-[4-(ACETYLAMINO)PHENYL]CYCLOPROPANECARBOXAMIDE | 4lvb | A | H191 S275 | 20N | | N-[4-(ACETYLAMINO)PHENYL]CYCLOPROPANECARBOXAMIDE | 4lvb | B | H191 S275 | 20P | | (1S,2S)-2-PHENYL-N-(PYRIDIN-4-YL) CYCLOPROPANECARBOXAMIDE | 4lvf | A | H191 S275 | 20P | | (1S,2S)-2-PHENYL-N-(PYRIDIN-4-YL) CYCLOPROPANECARBOXAMIDE | 4lvf | B | H191 S275 | 20O | | (1S,2S)-N-[4-(PHENYLSULFONYL)PHENYL]-2-(PYRIDIN-3-YL) CYCLOPROPANECARBOXAMIDE | 4lvg | A | H191 S275 | 20O | | (1S,2S)-N-[4-(PHENYLSULFONYL)PHENYL]-2-(PYRIDIN-3-YL) CYCLOPROPANECARBOXAMIDE | 4lvg | B | H191 S275 | 20R | | N-[4-(PHENYLSULFONYL)BENZYL]-2H-PYRAZOLO[3,4- B]PYRIDINE-5-CARBOXAMIDE | 4m6p | A | H191 S275 | 20R | | N-[4-(PHENYLSULFONYL)BENZYL]-2H-PYRAZOLO[3,4- B]PYRIDINE-5-CARBOXAMIDE | 4m6p | B | H191 S275 | 20T | | 1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-N-(4-{[3- (TRIFLUOROMETHYL)PHENYL]SULFONYL}BENZYL)-1H- PYRAZOLO[3,4-B]PYRIDINE-5-CARBOXAMIDE | 4m6q | A | H191 S275 | 20T | | 1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-N-(4-{[3- (TRIFLUOROMETHYL)PHENYL]SULFONYL}BENZYL)-1H- PYRAZOLO[3,4-B]PYRIDINE-5-CARBOXAMIDE | 4m6q | B | H191 S275 | LTS | | 2-CYANO-1-PYRIDIN-4-YL-3-(4-{[3-(TRIFLUOROMETHOXY) PHENYL]SULFONYL}BENZYL)GUANIDINE | 4lts | A | H191 S275 | LTS | | 2-CYANO-1-PYRIDIN-4-YL-3-(4-{[3-(TRIFLUOROMETHOXY) PHENYL]SULFONYL}BENZYL)GUANIDINE | 4lts | B | H191 S275 | LWW | | N-(4-(PHENYLSULFONYL)BENZYL)-1H-PYRROLO[3,2-C]PYRIDINE- 2-CARBOXAMIDE | 4lww | A | H191 S275 | LWW | | N-(4-(PHENYLSULFONYL)BENZYL)-1H-PYRROLO[3,2-C]PYRIDINE- 2-CARBOXAMIDE | 4lww | B | H191 S275 | 2ZM | | 5-NITRO-1H-BENZIMIDAZOLE | 4n9c | A | H191 S275 | 2ZM | | 5-NITRO-1H-BENZIMIDAZOLE | 4n9c | B | H191 S275 | 2HJ | | 4-({[(4-TERT-BUTYLPHENYL)SULFONYL]AMINO}METHYL)-N- (PYRIDIN-3-YL)BENZAMIDE | 4n9d | A | H191 S275 | 2HJ | | 4-({[(4-TERT-BUTYLPHENYL)SULFONYL]AMINO}METHYL)-N- (PYRIDIN-3-YL)BENZAMIDE | 4n9d | B | H191 S275 | 2HL | | 1-[(1-BENZOYLPIPERIDIN-4-YL)METHYL]-N-(PYRIDIN-3-YL)- 1H-BENZIMIDAZOLE-5-CARBOXAMIDE | 4n9e | A | H191 S275 | 2HL | | 1-[(1-BENZOYLPIPERIDIN-4-YL)METHYL]-N-(PYRIDIN-3-YL)- 1H-BENZIMIDAZOLE-5-CARBOXAMIDE | 4n9e | B | H191 S275 | 1XC | | 6-({4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}CARBAMOYL)- 1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)IMIDAZO[1,2- A]PYRIDIN-1-IUM | 4l4l | A | H191 S275 | 1XC | | 6-({4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}CARBAMOYL)- 1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)IMIDAZO[1,2- A]PYRIDIN-1-IUM | 4l4l | B | H191 S275 | 1XD | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}IMIDAZO[1,2- A]PYRIDINE-7-CARBOXAMIDE | 4l4m | A | H191 S275 | 1XD | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}IMIDAZO[1,2- A]PYRIDINE-7-CARBOXAMIDE | 4l4m | B | H191 S275 | 2RM | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}INDOLIZINE-6- CARBOXAMIDE | 4o0z | A | H191 S275 | 2RM | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}INDOLIZINE-6- CARBOXAMIDE | 4o0z | B | H191 S275 | 2QF | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}INDOLIZINE-7- CARBOXAMIDE | 4o10 | A | H191 S275 | 2QF | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}INDOLIZINE-7- CARBOXAMIDE | 4o10 | B | H191 S275 | 2QG | | 2-[6-(4-CHLOROPHENOXY)HEXYL]-1-CYANO-3-PYRIDIN-4- YLGUANIDINE | 4o12 | A | H191 S275 | 2QG | | 2-[6-(4-CHLOROPHENOXY)HEXYL]-1-CYANO-3-PYRIDIN-4- YLGUANIDINE | 4o12 | B | H191 S275 | 2P1 | | N-(4-{[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL}BENZYL)-2H- PYRAZOLO[3,4-B]PYRIDINE-5-CARBOXAMIDE | 4o13 | A | H191 S275 | 2P1 | | N-(4-{[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL}BENZYL)-2H- PYRAZOLO[3,4-B]PYRIDINE-5-CARBOXAMIDE | 4o13 | B | H191 S275 | 2P1 | | N-(4-{[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL}BENZYL)-2H- PYRAZOLO[3,4-B]PYRIDINE-5-CARBOXAMIDE | 4o15 | A | H191 S275 | 2P1 | | N-(4-{[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL}BENZYL)-2H- PYRAZOLO[3,4-B]PYRIDINE-5-CARBOXAMIDE | 4o15 | B | H191 S275 | 1XC | | 6-({4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}CARBAMOYL)- 1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)IMIDAZO[1,2- A]PYRIDIN-1-IUM | 4o16 | A | H191 S275 | 1QS | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}IMIDAZO[1,2- A]PYRIDINE-6-CARBOXAMIDE | 4o28 | A | H191 S275 | 1QS | | N-{4-[(3,5-DIFLUOROPHENYL)SULFONYL]BENZYL}IMIDAZO[1,2- A]PYRIDINE-6-CARBOXAMIDE | 4o28 | B | H191 S275 | 3TQ | | N-(4-{(S)-[1-(2-METHYLPROPYL)PIPERIDIN-4- YL]SULFINYL}BENZYL)FURO[2,3-C]PYRIDINE-2-CARBOXAMIDE | 4wq6 | A | H191 S275 | 3TQ | | N-(4-{(S)-[1-(2-METHYLPROPYL)PIPERIDIN-4- YL]SULFINYL}BENZYL)FURO[2,3-C]PYRIDINE-2-CARBOXAMIDE | 4wq6 | B | H191 S275 | BEF | | TRIFLUOROBERYLLATE(1-) | 3dhf | A | K415 | BEF | | TRIFLUOROBERYLLATE(1-) | 3dhf | B | K415 | BEF | | TRIFLUOROBERYLLATE(1-) | 3dkl | A | K415 | BEF | | TRIFLUOROBERYLLATE(1-) | 3dkl | B | K415 | POP | | DIPHOSPHATE(2-) | 3dhd | A | K415 K423 | POP | | DIPHOSPHATE(2-) | 3dhd | B | K415 K423 | POP | | DIPHOSPHATE(2-) | 3dhf | A | K415 K423 | POP | | DIPHOSPHATE(2-) | 3dhf | B | K415 K423 | PRP | | 5-O-PHOSPHONO-ALPHA-D-RIBOFURANOSYL DIPHOSPHATE | 3dkl | A | K415 K423 | PRP | | 5-O-PHOSPHONO-ALPHA-D-RIBOFURANOSYL DIPHOSPHATE | 3dkl | B | K415 K423 | PRP | | 5-O-PHOSPHONO-ALPHA-D-RIBOFURANOSYL DIPHOSPHATE | 3dkj | A | K423 | PRP | | 5-O-PHOSPHONO-ALPHA-D-RIBOFURANOSYL DIPHOSPHATE | 3dkj | B | K423 | POP | | DIPHOSPHATE(2-) | 4m6q | B | K423 | DGB | | N-[4-(1-BENZOYLPIPERIDIN-4-YL)BUTYL]-3-PYRIDIN-3-YLPROPANAMIDE | 2gvj | B | S275 | 20J | | 7-CHLORO-3-METHYL-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE | 4lv9 | A | S275 | 2HH | | 1-METHYL-N-(PYRIDIN-3-YL)-1H-PYRAZOLE-5-CARBOXAMIDE | 4n9b | A | S275 | 2HH | | 1-METHYL-N-(PYRIDIN-3-YL)-1H-PYRAZOLE-5-CARBOXAMIDE | 4n9b | B | S275 | DGB | | N-[4-(1-BENZOYLPIPERIDIN-4-YL)BUTYL]-3-PYRIDIN-3-YLPROPANAMIDE | 4o1d | A | S275 | DGB | | N-[4-(1-BENZOYLPIPERIDIN-4-YL)BUTYL]-3-PYRIDIN-3-YLPROPANAMIDE | 4o1d | B | S275 |
| LBS | AA sequence | # species | Species |
A244 | GYSVPAAEHST | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | A245 | YSVPAAEHSTI | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | A379 | SIENVSFGSGG | 2 | Mus musculus, Rattus norvegicus | A379 | SIENIAFGSGG | 1 | Homo sapiens | D16 | ILLATDSYKVT | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | D219 | NFKGTDTVAGI | 2 | Mus musculus, Rattus norvegicus | D219 | NFKGTDTVAGL | 1 | Homo sapiens | D313 | LIIRPDSGNPL | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | D354 | RVIQGDGVDIN | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | E246 | SVPAAEHSTIT | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | E376 | KKWSIENVSFG | 2 | Mus musculus, Rattus norvegicus | E376 | KMWSIENIAFG | 1 | Homo sapiens | F193 | YKLHDFGYRGV | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | G185 | SGNLDGLEYKL | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | G217 | LVNFKGTDTVA | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | G353 | LRVIQGDGVDI | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | G383 | VSFGSGGALLQ | 2 | Mus musculus, Rattus norvegicus | G383 | IAFGSGGGLLQ | 1 | Homo sapiens | G384 | SFGSGGALLQK | 2 | Mus musculus, Rattus norvegicus | G384 | AFGSGGGLLQK | 1 | Homo sapiens | H191 | LEYKLHDFGYR | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | H247 | VPAAEHSTITA | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | I309 | TEAPLIIRPDS | 2 | Mus musculus, Rattus norvegicus | I309 | TQAPLIIRPDS | 1 | Homo sapiens | I351 | PYLRVIQGDGV | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | I378 | WSIENVSFGSG | 2 | Mus musculus, Rattus norvegicus | I378 | WSIENIAFGSG | 1 | Homo sapiens | K189 | DGLEYKLHDFG | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | K400 | LNCSFKCSYVV | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | K415 | GVNVFKDPVAD | 2 | Mus musculus, Rattus norvegicus | K415 | GINVFKDPVAD | 1 | Homo sapiens | K423 | VADPNKRSKKG | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | N377 | KWSIENVSFGS | 2 | Mus musculus, Rattus norvegicus | N377 | MWSIENIAFGS | 1 | Homo sapiens | P273 | QFSSVPVSVVS | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | P307 | RSTEAPLIIRP | 2 | Mus musculus, Rattus norvegicus | P307 | RSTQAPLIIRP | 1 | Homo sapiens | Q305 | VSRSTEAPLII | 2 | Mus musculus, Rattus norvegicus | Q305 | VSRSTQAPLII | 1 | Homo sapiens | R196 | HDFGYRGVSSQ | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | R311 | APLIIRPDSGN | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | R349 | LPPYLRVIQGD | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | R392 | LQKLTRDLLNC | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | S241 | PVPGYSVPAAE | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | S275 | SSVPVSVVSDS | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | S398 | DLLNCSFKCSY | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | T304 | IVSRSTEAPLI | 2 | Mus musculus, Rattus norvegicus | T304 | IVSRSTQAPLI | 1 | Homo sapiens | V242 | VPGYSVPAAEH | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | V350 | PPYLRVIQGDG | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | Y18 | LATDSYKVTHY | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | Y188 | LDGLEYKLHDF | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | Y240 | DPVPGYSVPAA | 3 | Homo sapiens, Mus musculus, Rattus norvegicus |
 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Lollipop-style diagram of mutations at LBS in amino-acid sequence.
Lollipop-style diagram of mutations at LBS in amino-acid sequence. 
 Cancer type specific mutLBS sorted by frequency
Cancer type specific mutLBS sorted by frequency Relative protein structure stability change (ΔΔE) using Mupro 1.1
Relative protein structure stability change (ΔΔE) using Mupro 1.1  : nsSNV at non-LBS
: nsSNV at non-LBS : nsSNV at LBS
: nsSNV at LBS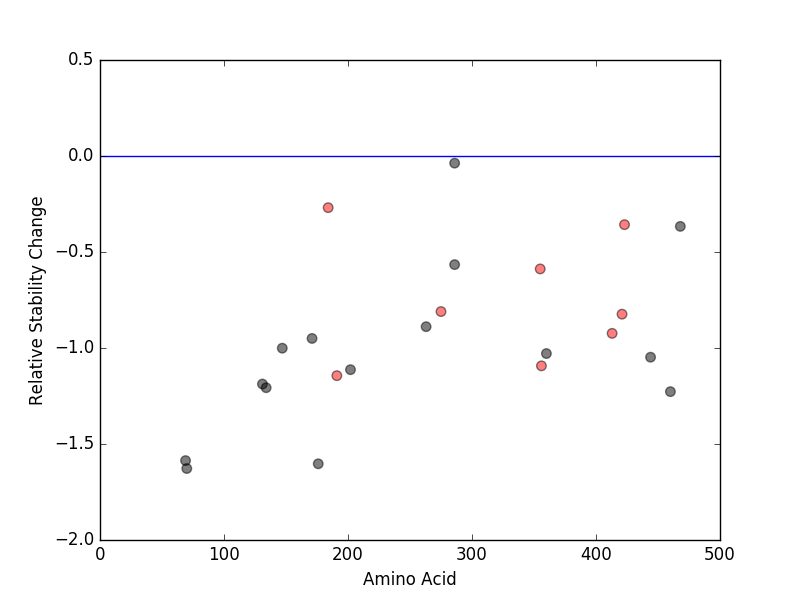
 nsSNVs sorted by the relative stability change of protein structure by each mutation
nsSNVs sorted by the relative stability change of protein structure by each mutation  Structure image for NAMPT from PDB
Structure image for NAMPT from PDB Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types
Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types Differential co-expressed gene network based on protein-protein interaction data (CePIN)
Differential co-expressed gene network based on protein-protein interaction data (CePIN) Gene level disease information (DisGeNet)
Gene level disease information (DisGeNet)  Mutation level pathogenic information (ClinVar annotation)
Mutation level pathogenic information (ClinVar annotation)  Gene expression profile of anticancer drug treated cell-lines (CCLE)
Gene expression profile of anticancer drug treated cell-lines (CCLE)
 Gene-centered drug-gene interaction network
Gene-centered drug-gene interaction network 
 Drug information targeting mutLBSgene (Approved drugs only)
Drug information targeting mutLBSgene (Approved drugs only)

 Gene-centered ligand-gene interaction network
Gene-centered ligand-gene interaction network 
 Ligands binding to mutated ligand binding site of NAMPT go to BioLip
Ligands binding to mutated ligand binding site of NAMPT go to BioLip Multiple alignments for P43490 in multiple species
Multiple alignments for P43490 in multiple species 
