|
mutLBSgeneDB |
| |
| |
| |
| |
| |
| |
|
| Gene summary for CYP2C9 |
 Gene summary Gene summary |
| Basic gene Info. | Gene symbol | CYP2C9 |
| Gene name | cytochrome P450, family 2, subfamily C, polypeptide 9 | |
| Synonyms | CPC9|CYP2C|CYP2C10|CYPIIC9|P450IIC9 | |
| Cytomap | UCSC genome browser: 10q24 | |
| Type of gene | protein-coding | |
| RefGenes | NM_000771.3, | |
| Description | cytochrome P-450 S-mephenytoin 4-hydroxylasecytochrome P-450MPcytochrome P450 2C9cytochrome P450 PB-1flavoprotein-linked monooxygenasemicrosomal monooxygenasexenobiotic monooxygenase | |
| Modification date | 20141211 | |
| dbXrefs | MIM : 601130 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000138109 | ||
| HPRD : 03084 | ||
| Vega : OTTHUMG00000018805 | ||
| Protein | UniProt: P11712 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_CYP2C9 | |
| BioGPS: 1559 | ||
| Pathway | NCI Pathway Interaction Database: CYP2C9 | |
| KEGG: CYP2C9 | ||
| REACTOME: CYP2C9 | ||
| Pathway Commons: CYP2C9 | ||
| Context | iHOP: CYP2C9 | |
| ligand binding site mutation search in PubMed: CYP2C9 | ||
| UCL Cancer Institute: CYP2C9 | ||
| Assigned class in mutLBSgeneDB | A: This gene has a literature evidence and it belongs to targetable_mutLBSgenes. | |
| References showing study about ligand binding site mutation for CYP2C9. | 1. "Haining RL, Jones JP, Henne KR, Fisher MB, Koop DR, Trager WF, Rettie AE. Enzymatic determinants of the substrate specificity of CYP2C9: role of B'-C loop residues in providing the pi-stacking anchor site for warfarin binding. Biochemistry. 1999 Mar 16;38(11):3285-92. PubMed PMID: 10079071. " 10079071 2. "Dickmann LJ, Locuson CW, Jones JP, Rettie AE. Differential roles of Arg97, Asp293, and Arg108 in enzyme stability and substrate specificity of CYP2C9. Mol Pharmacol. 2004 Apr;65(4):842-50. PubMed PMID: 15044613. " 15044613 | |
 Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez |
| GO ID | GO Term | PubMed ID | GO:0016098 | monoterpenoid metabolic process | 16401082 | GO:0017144 | drug metabolic process | 19219744 | GO:0019627 | urea metabolic process | 19029318 | GO:0032787 | monocarboxylic acid metabolic process | 19651758 | GO:0042738 | exogenous drug catabolic process | 18619574 | GO:0043603 | cellular amide metabolic process | 19651758 | GO:0055114 | oxidation-reduction process | 16401082 | GO:0070989 | oxidative demethylation | 18619574 |
| Top |
| Ligand binding site mutations for CYP2C9 |
 Cancer type specific mutLBS sorted by frequency Cancer type specific mutLBS sorted by frequency |
| LBS | AAchange of nsSNV | Cancer type | # samples | E444 | E444G | COAD | 2 | S429,R433 | G431E | SKCM | 2 | L233 | N231K | COAD | 1 | R433 | R433W | COAD | 1 | E444 | E444K | COAD | 1 | N107 | A106V | KIRC | 1 | L102,F100 | P101Q | LUAD | 1 | A103 | A103V | LUAD | 1 | L391 | I389T | LUSC | 1 | E444 | E444D | OV | 1 | R108 | G109R | SKCM | 1 | L362,T364 | P363S | SKCM | 1 | R124,W120 | E122K | SKCM | 1 | L391 | S390F | SKCM | 1 |
| cf) Cancer type abbreviation. BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, COAD: Colon adenocarcinoma, GBM: Glioblastoma multiforme, LGG: Brain lower grade glioma, HNSC: Head and neck squamous cell carcinoma, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute myeloid leukemia, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PRAD: Prostate adenocarcinoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, THCA: Thyroid carcinoma, UCEC: Uterine corpus endometrial carcinoma. |
| Top |
| Protein structure related information for CYP2C9 |
 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Mupro score denotes assessment of the effect of mutations on thermodynamic stability. (ΔΔE<0: mutation decreases stability, ΔΔE>0: mutation increases stability) |
 : nsSNV at non-LBS : nsSNV at non-LBS : nsSNV at LBS : nsSNV at LBS |
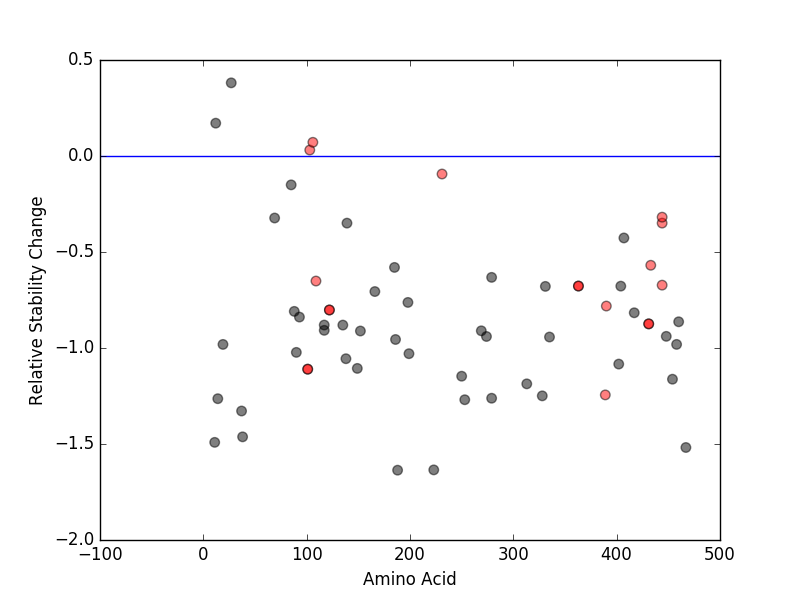 |
 nsSNVs sorted by the relative stability change of protein structure by each mutation nsSNVs sorted by the relative stability change of protein structure by each mutation Blue: mutations of positive stability change. and red : the most recurrent mutation for this gene. |
| LBS | AAchange of nsSNV | Relative stability change | N107 | A106V | 0.071331411 | A103 | A103V | 0.031067281 | L391 | I389T | -1.2443179 | F100 | P101Q | -1.1103241 | L102 | P101Q | -1.1103241 | S429 | G431E | -0.87435855 | R433 | G431E | -0.87435855 | W120 | E122K | -0.80202708 | R124 | E122K | -0.80202708 | L391 | S390F | -0.78152168 | L362 | P363S | -0.6773486 | T364 | P363S | -0.6773486 | E444 | E444G | -0.67260317 | R108 | G109R | -0.65122539 | R433 | R433W | -0.56910267 | E444 | E444D | -0.34949731 | E444 | E444K | -0.31850736 | L233 | N231K | -0.093827882 |
| (MuPro1.1: Jianlin Cheng et al., Prediction of Protein Stability Changes for Single-Site Mutations Using Support Vector Machines, PROTEINS: Structure, Function, and Bioinformatics. 2006, 62:1125-1132) |
 Structure image for CYP2C9 from PDB Structure image for CYP2C9 from PDB |
| PDB ID | PDB title | PDB structure | 1OG2 | STRUCTURE OF HUMAN CYTOCHROME P450 CYP2C9 |  |
| Top |
| Differential gene expression and gene-gene network for CYP2C9 |
 Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types |
 Differential co-expressed gene network based on protein-protein interaction data (CePIN) Differential co-expressed gene network based on protein-protein interaction data (CePIN) |
| Top |
| Top |
| Phenotype information for CYP2C9 |
 Gene level disease information (DisGeNet) Gene level disease information (DisGeNet) |
| Disease ID | Disease name | # PubMed | Association type |
| umls:C0019080 | Hemorrhage | 13 | Biomarker, GeneticVariation |
| umls:C0860207 | Drug-Induced Liver Injury | 5 | Biomarker, GeneticVariation |
| umls:C0013182 | Drug Hypersensitivity | 4 | Biomarker, GeneticVariation |
| umls:C0750384 | Coumarin Resistance | 2 | Biomarker, GeneticVariation |
| umls:C3658338 | Drug-Related Side Effects and Adverse Reactions | 2 | Biomarker |
| umls:C0030922 | Peptic Ulcer Hemorrhage | 1 | Biomarker, GeneticVariation |
| umls:C0006118 | Brain Neoplasms | 1 | Biomarker |
| umls:C2609414 | Acute Kidney Injury | 1 | Biomarker |
| umls:C0878544 | Cardiomyopathies | 1 | Biomarker |
| umls:C0027707 | Nephritis, Interstitial | 1 | Biomarker |
| umls:C0027765 | Nervous System Diseases | 1 | Biomarker |
 Mutation level pathogenic information (ClinVar annotation) Mutation level pathogenic information (ClinVar annotation) |
| Allele ID | AA change | Clinical significance | Origin | Phenotype IDs |
| Top |
| Pharmacological information for CYP2C9 |
 Gene expression profile of anticancer drug treated cell-lines (CCLE) Gene expression profile of anticancer drug treated cell-lines (CCLE)Heatmap showing the correlation between gene expression and drug response across all the cell-lines. We chose the top 20 among 138 drugs.We used Pearson's correlation coefficient. |
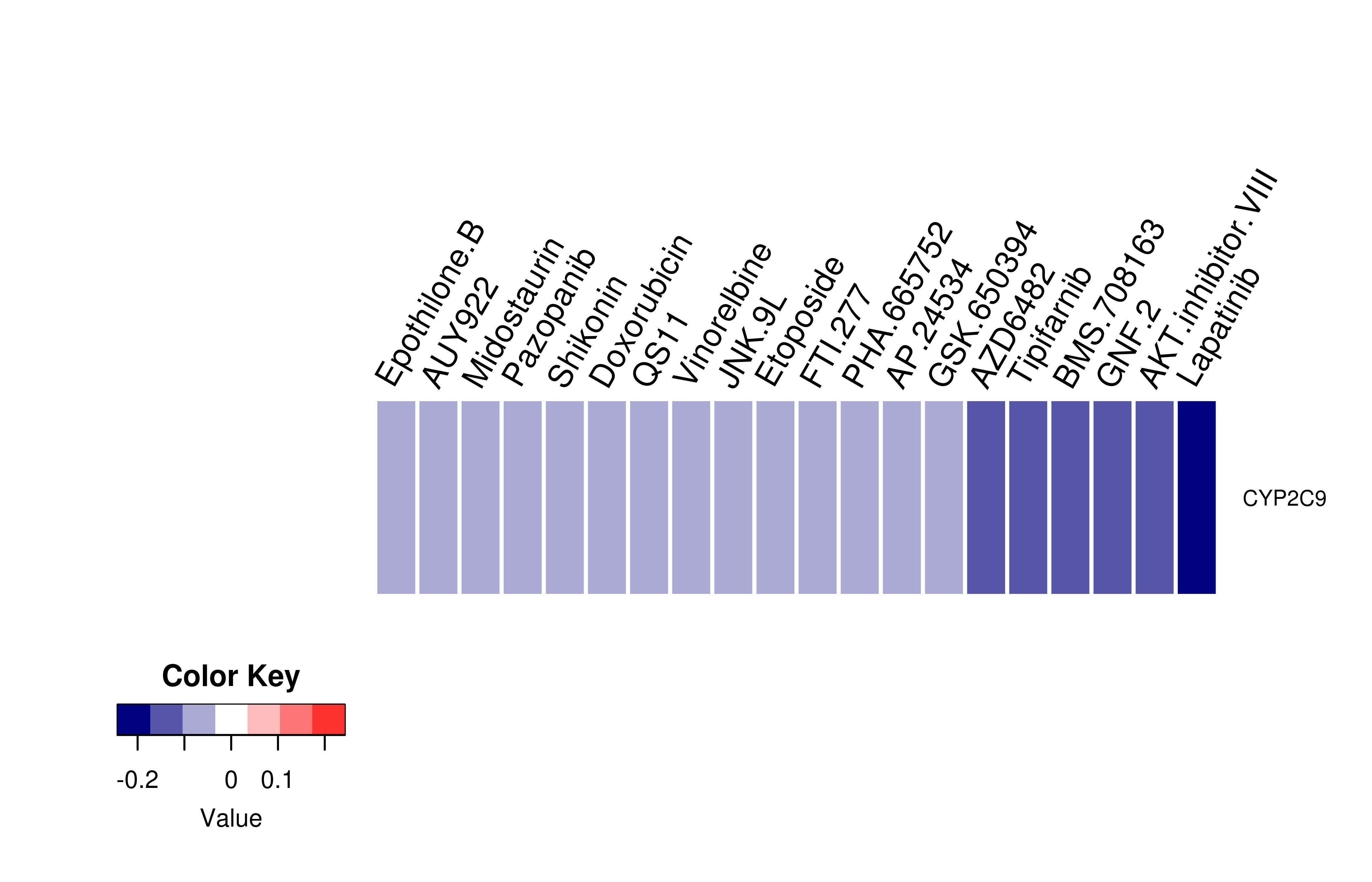 |
 Gene-centered drug-gene interaction network Gene-centered drug-gene interaction network |
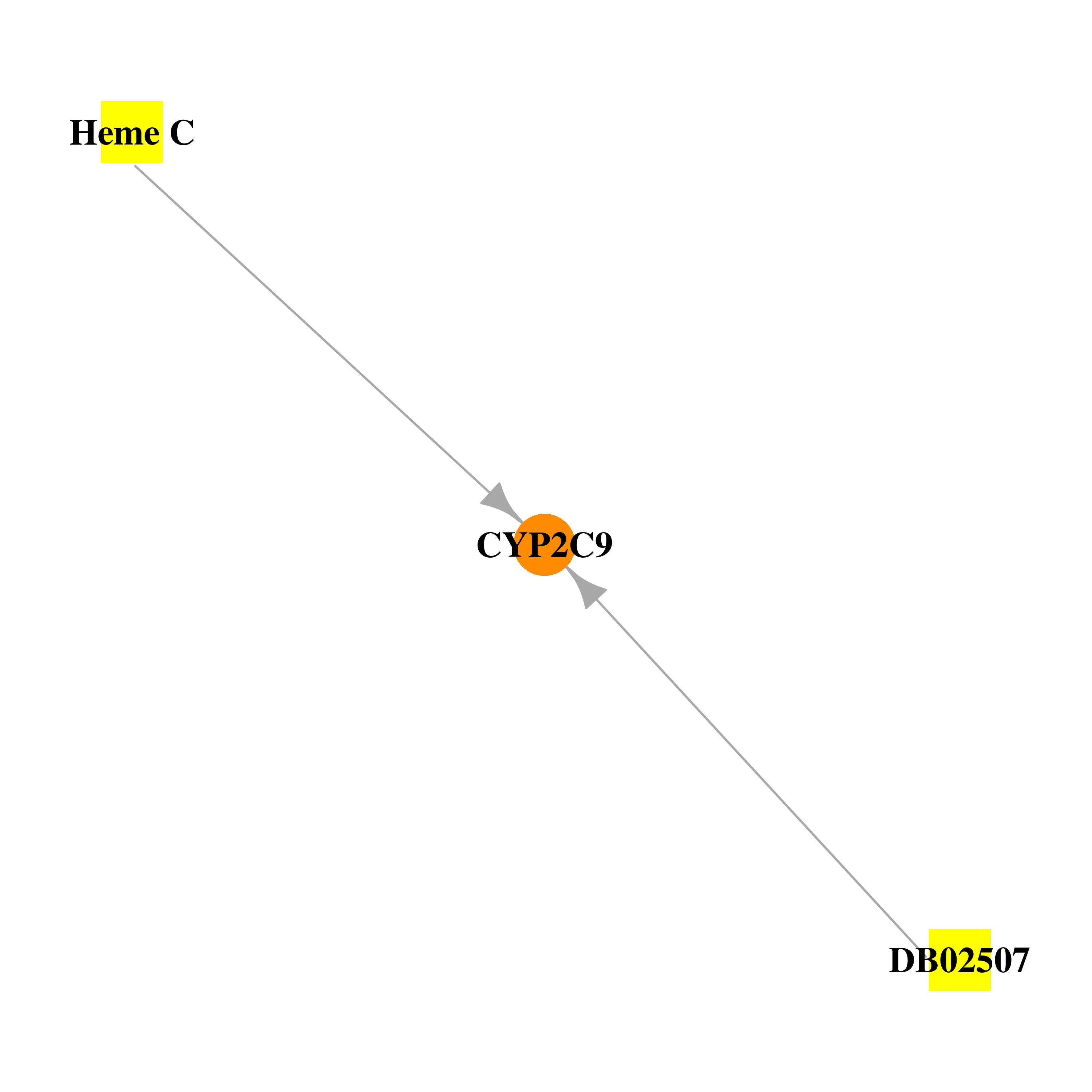 |
 Drug information targeting mutLBSgene (Approved drugs only) Drug information targeting mutLBSgene (Approved drugs only) |
| Drug status | DrugBank ID | Name | Type | Drug structure |
| Experimental | DB02507 | 4-Hydroxy-3-[(1s)-3-Oxo-1-Phenylbutyl]-2h-Chromen-2-One | Small molecule | 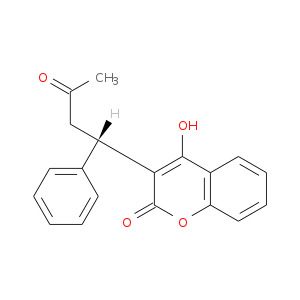 |
| Experimental | DB03317 | Heme C | Small molecule | 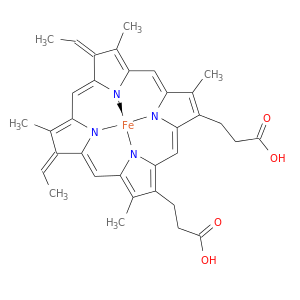 |
 Gene-centered ligand-gene interaction network Gene-centered ligand-gene interaction network |
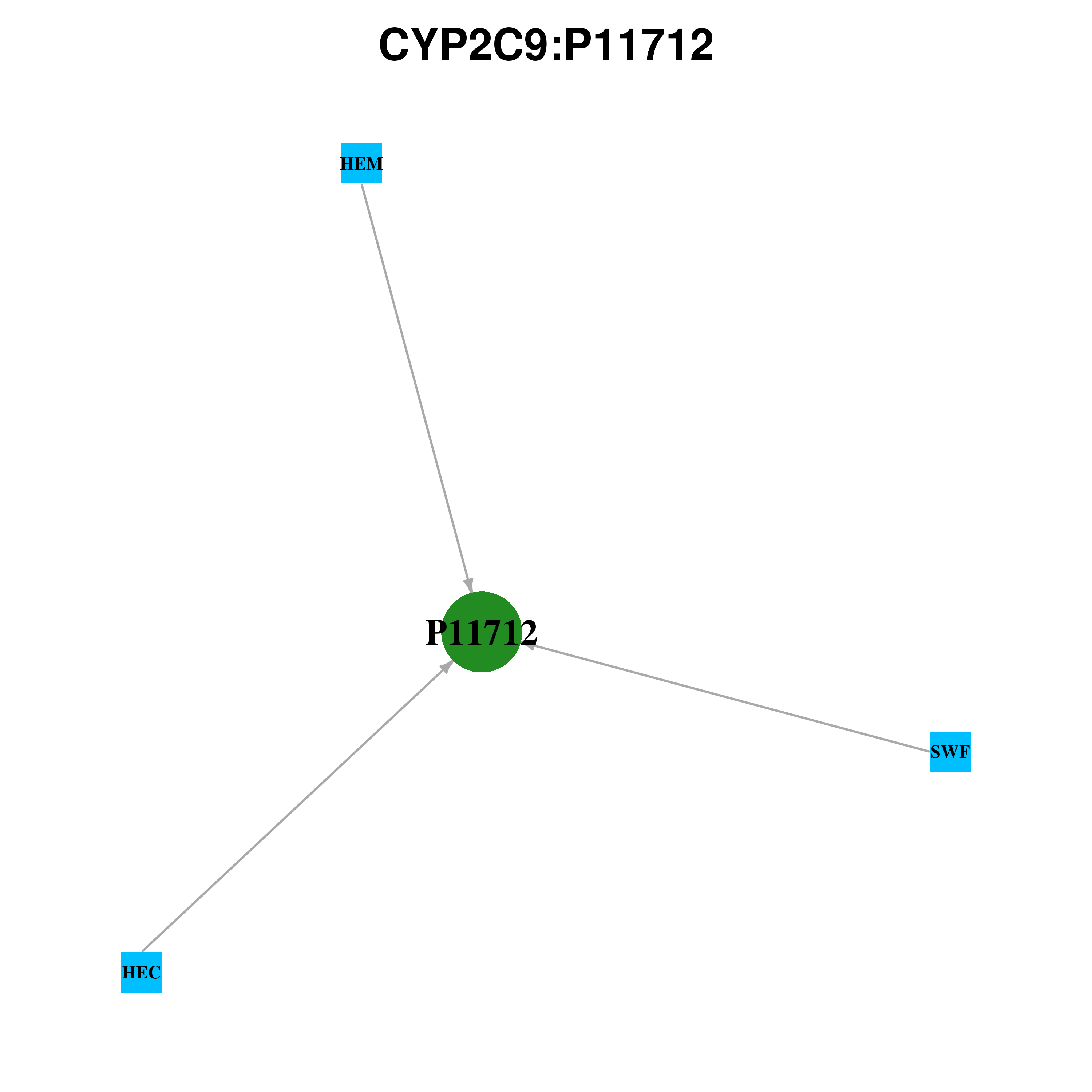 |
 Ligands binding to mutated ligand binding site of CYP2C9 go to BioLip Ligands binding to mutated ligand binding site of CYP2C9 go to BioLip |
| Ligand ID | Ligand short name | Ligand long name | PDB ID | PDB name | mutLBS | SWF | S-WARFARIN | 1og5 | A | F100 L102 A103 T364 | SWF | S-WARFARIN | 1og5 | B | F100 L102 A103 T364 | 2QJ | (2R)-N-{4-[(3-BROMOPHENYL)SULFONYL]-2-CHLOROPHENYL}-3, 3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANAMIDE | 4nz2 | A | N107 L233 L362 | 2QJ | (2R)-N-{4-[(3-BROMOPHENYL)SULFONYL]-2-CHLOROPHENYL}-3, 3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANAMIDE | 4nz2 | B | N107 L362 | FLP | FLURBIPROFEN | 1r9o | A | R108 | HEM | HEME B | 1r9o | A | W120 L362 S429 R433 E444 | HEC | FERROHEME C(2-) | 1og2 | B | W120 R124 L362 L391 S429 R433 | HEC | FERROHEME C(2-) | 1og5 | B | W120 R124 L362 L391 S429 R433 E444 | HEC | FERROHEME C(2-) | 1og5 | A | W120 R124 L362 S429 R433 | HEM | HEME B | 4nz2 | A | W120 R124 L362 S429 R433 | HEM | HEME B | 4nz2 | B | W120 R124 L362 S429 R433 | HEC | FERROHEME C(2-) | 1og2 | A | W120 R124 L362 S429 R433 E444 |
| Top |
| Conservation information for LBS of CYP2C9 |
 Multiple alignments for P11712 in multiple species Multiple alignments for P11712 in multiple species |
| LBS | AA sequence | # species | Species |
 |
Copyright © 2016-Present - The University of Texas Health Science Center at Houston |
