| Basic gene Info. | Gene symbol | ERBB2 |
| Gene name | erb-b2 receptor tyrosine kinase 2 |
| Synonyms | CD340|HER-2|HER-2/neu|HER2|MLN 19|NEU|NGL|TKR1 |
| Cytomap | UCSC genome browser: 17q12 |
| Type of gene | protein-coding |
| RefGenes | NM_001005862.2,
NM_001289936.1,NM_001289937.1,NM_001289938.1,NM_004448.3,
NR_110535.1, |
| Description | c-erb B2/neu proteinherstatinhuman epidermal growth factor receptor 2metastatic lymph node gene 19 proteinneuro/glioblastoma derived oncogene homologneuroblastoma/glioblastoma derived oncogene homologp185erbB2proto-oncogene Neuproto-oncogene c-Erb |
| Modification date | 20141221 |
| dbXrefs | MIM : 164870 |
| HGNC : HGNC |
| Ensembl : ENSG00000141736 |
| HPRD : 01281 |
| Vega : OTTHUMG00000179300 |
| Protein | UniProt: P04626
go to UniProt's Cross Reference DB Table |
| Expression | CleanEX: HS_ERBB2 |
| BioGPS: 2064 |
| Pathway | NCI Pathway Interaction Database: ERBB2 |
| KEGG: ERBB2 |
| REACTOME: ERBB2 |
| Pathway Commons: ERBB2 |
| Context | iHOP: ERBB2 |
| ligand binding site mutation search in PubMed: ERBB2 |
| UCL Cancer Institute: ERBB2 |
| Assigned class in mutLBSgeneDB | A: This gene has a literature evidence and it belongs to targetable_mutLBSgenes. |
| References showing study about ligand binding site mutation for ERBB2. | 1. "Fan YX, Wong L, Ding J, Spiridonov NA, Johnson RC, Johnson GR. Mutational activation of ErbB2 reveals a new protein kinase autoinhibition mechanism. J Biol Chem. 2008 Jan 18;283(3):1588-96. Epub 2007 Nov 26. PubMed PMID: 18039657. " 18039657
|
| cf) Cancer type abbreviation. BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, COAD: Colon adenocarcinoma, GBM: Glioblastoma multiforme, LGG: Brain lower grade glioma, HNSC: Head and neck squamous cell carcinoma, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute myeloid leukemia, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PRAD: Prostate adenocarcinoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, THCA: Thyroid carcinoma, UCEC: Uterine corpus endometrial carcinoma. |
| HER2 (ERBB2) belongs to a family of receptor tyrosine kinases (RTKs) that includes EGFR/ERBB1, HER2/ERBB2/NEU, HER3/ERBB3, and HER4/ERBB4. The gene for HER2 is located on chromosome 17 and has been found to be amplified with an increased copy number in several cancers (Jorgensen 2010). Amplification of HER2 has been found to promote tumorigenesis and to be involved in the pathogenesis of several human cancers (Moasser 2007).To date, no ligand has been identified for HER2. However, HER2 appears to be the preferential dimerization partner for all members of the ERBB family (Graus-Porta et al. 1997). The binding of ligand followed by HER2 hetero-dimerization results in activation of HER2 tyrosine kinase activity. Activated HER2 then phosphorylates its substrates, leading to activation of multiple downstream pathways within the cell, including the PI3K-AKT-mTOR pathway, which is involved in cell survival, and the RAS-RAF-MEK-ERK pathway, which is involved in cell proliferation. Lovly, C.M., Shi, C., Watson, G.T., Horn, L., Pohlmann, P., Goff, L.W. 2015. HER2 (ERBB2). My Cancer Genome https://www.mycancergenome.org/content/gene/erbb2/ (Updated December 2015). |
| Disease ID | Disease name | # PubMed | Association type |
| umls:C1458155 | Breast Neoplasms | 508 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification, Therapeutic |
| umls:C0027627 | Neoplasm Metastasis | 334 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0001418 | Adenocarcinoma | 202 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0007131 | Carcinoma, Non-Small-Cell Lung | 87 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0919267 | Ovarian Neoplasms | 54 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0033578 | Prostatic Neoplasms | 24 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0024121 | Lung Neoplasms | 21 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0152013 | Adenocarcinoma of lung | 21 | Biomarker, GeneticVariation |
| umls:C0014859 | Esophageal Neoplasms | 17 | Biomarker, GeneticVariation |
| umls:C0038356 | Stomach Neoplasms | 15 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0007134 | Carcinoma, Renal Cell | 14 | Biomarker |
| umls:C0017638 | Glioma | 13 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0206698 | Cholangiocarcinoma | 12 | AlteredExpression, Biomarker |
| umls:C0025149 | Medulloblastoma | 6 | AlteredExpression, Biomarker |
| umls:C0009375 | Colonic Neoplasms | 5 | AlteredExpression, Biomarker |
| umls:C0024667 | Mammary Neoplasms, Animal | 4 | Biomarker, GeneticVariation |
| umls:C0030354 | Papilloma | 4 | AlteredExpression, Biomarker |
| umls:C0024232 | Lymphatic Metastasis | 3 | Biomarker |
| umls:C2931822 | Nasopharyngeal carcinoma | 3 | Biomarker |
| umls:C0024668 | Mammary Neoplasms, Experimental | 2 | Biomarker |
| umls:C0027659 | Neoplasms, Experimental | 2 | Biomarker, GeneticVariation |
| umls:C0017185 | Gastrointestinal Neoplasms | 2 | Biomarker |
| umls:C0242656 | Disease Progression | 1 | Biomarker |
| umls:C0016978 | Gallbladder Neoplasms | 1 | Biomarker |
| umls:C0027643 | Neoplasm Recurrence, Local | 1 | Biomarker |
| umls:C0031117 | Peripheral Nervous System Diseases | 1 | Biomarker |
| Ligand ID | Ligand short name | Ligand long name | PDB ID | PDB name | mutLBS |
03Q | | 2-{2-[4-({5-CHLORO-6-[3-(TRIFLUOROMETHYL)PHENOXY]PYRIDIN-3-YL}AMINO)-5H-PYRROLO[3,2-D]PYRIMIDIN-5-YL]ETHOXY}ETHANOL | 3pp0 | A | L726 V734 A751 K753 E770 M774 L796 Q799 T862 | 03Q | | 2-{2-[4-({5-CHLORO-6-[3-(TRIFLUOROMETHYL)PHENOXY]PYRIDIN-3-YL}AMINO)-5H-PYRROLO[3,2-D]PYRIMIDIN-5-YL]ETHOXY}ETHANOL | 3pp0 | B | L726 V734 A751 K753 E770 M774 L796 Q799 T862 F1004 | 03P | | TAK-285 | 3rcd | A | L726 V734 A751 K753 L796 Q799 T862 F1004 | 03P | | TAK-285 | 3rcd | C | L726 V734 A751 K753 R784 L796 Q799 T862 F1004 |
 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Lollipop-style diagram of mutations at LBS in amino-acid sequence.
Lollipop-style diagram of mutations at LBS in amino-acid sequence. 
 Cancer type specific mutLBS sorted by frequency
Cancer type specific mutLBS sorted by frequency Clinical information for ERBB2 from My Cancer Genome.
Clinical information for ERBB2 from My Cancer Genome.  Protein structure of wild type (WT) and mutant type (MT) of ERBB2
Protein structure of wild type (WT) and mutant type (MT) of ERBB2 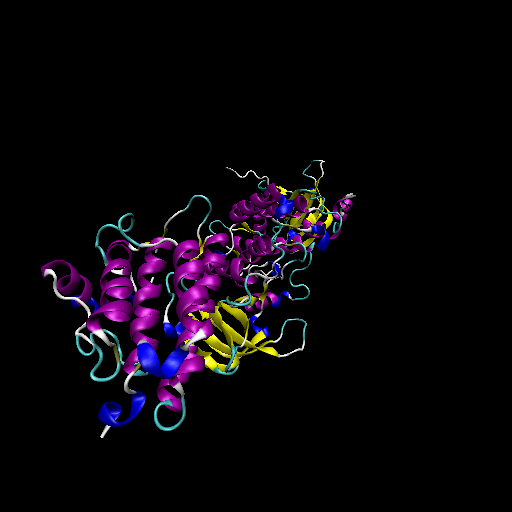
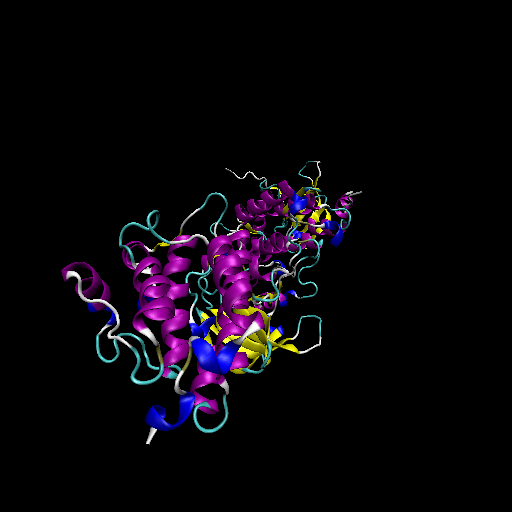
 Free energy of binding of drugs to wild type and mutant tpye of ERBB2
Free energy of binding of drugs to wild type and mutant tpye of ERBB2  Relative protein structure stability change (ΔΔE) using Mupro 1.1
Relative protein structure stability change (ΔΔE) using Mupro 1.1  : nsSNV at non-LBS
: nsSNV at non-LBS : nsSNV at LBS
: nsSNV at LBS
 nsSNVs sorted by the relative stability change of protein structure by each mutation
nsSNVs sorted by the relative stability change of protein structure by each mutation  Structure image for ERBB2 from PDB
Structure image for ERBB2 from PDB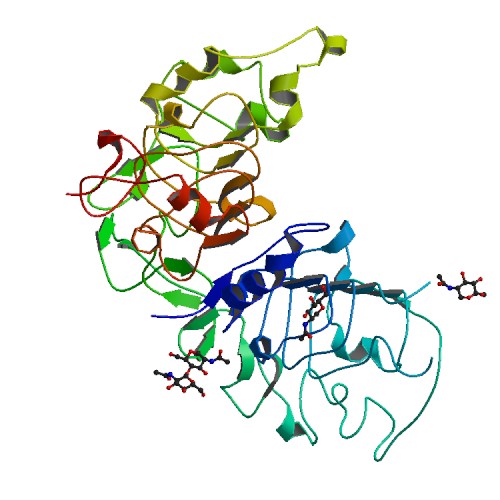
 Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types
Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types Differential co-expressed gene network based on protein-protein interaction data (CePIN)
Differential co-expressed gene network based on protein-protein interaction data (CePIN) Gene level disease information (DisGeNet)
Gene level disease information (DisGeNet)  Mutation level pathogenic information (ClinVar annotation)
Mutation level pathogenic information (ClinVar annotation)  Gene expression profile of anticancer drug treated cell-lines (CCLE)
Gene expression profile of anticancer drug treated cell-lines (CCLE)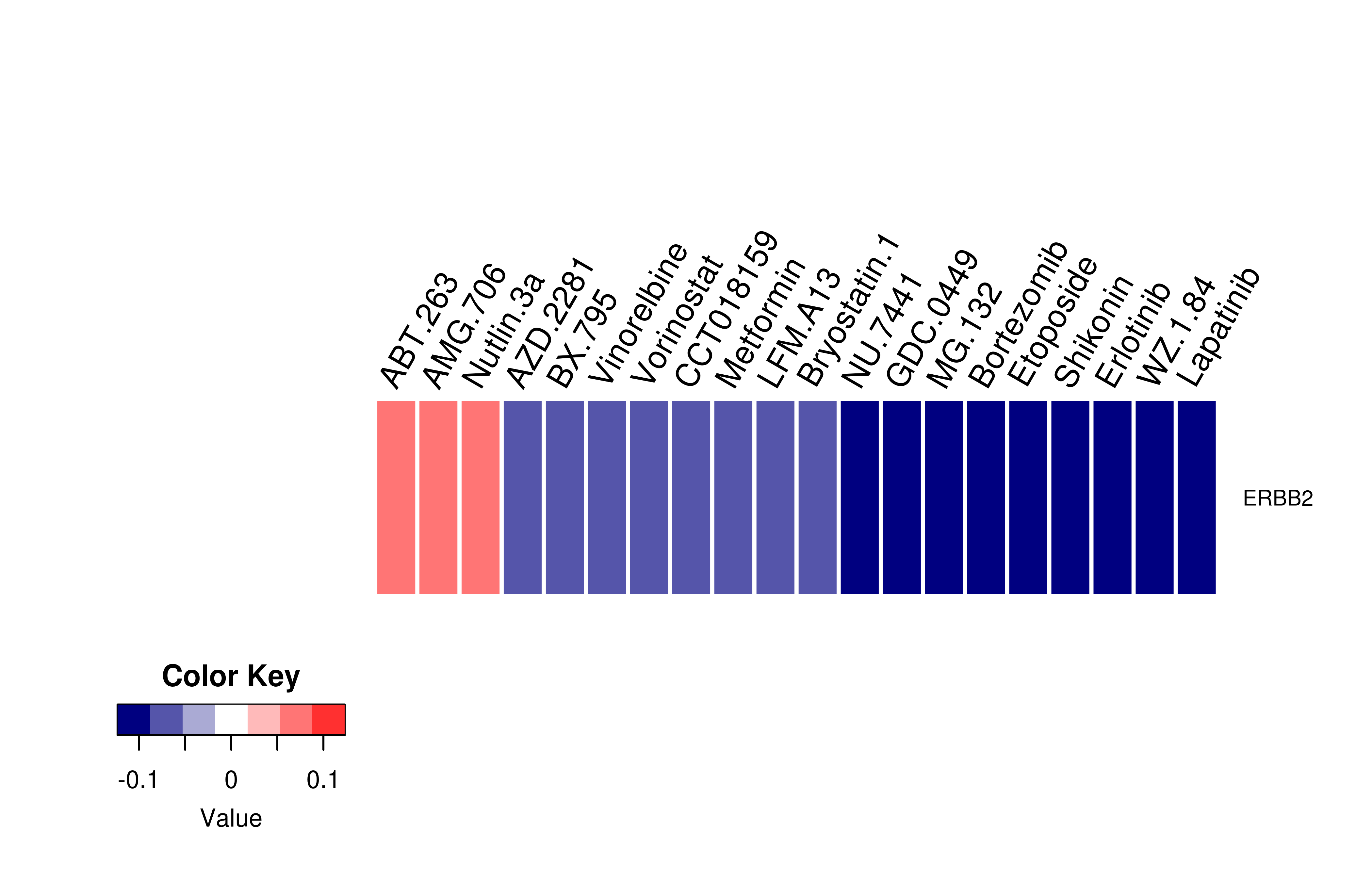
 Gene-centered drug-gene interaction network
Gene-centered drug-gene interaction network 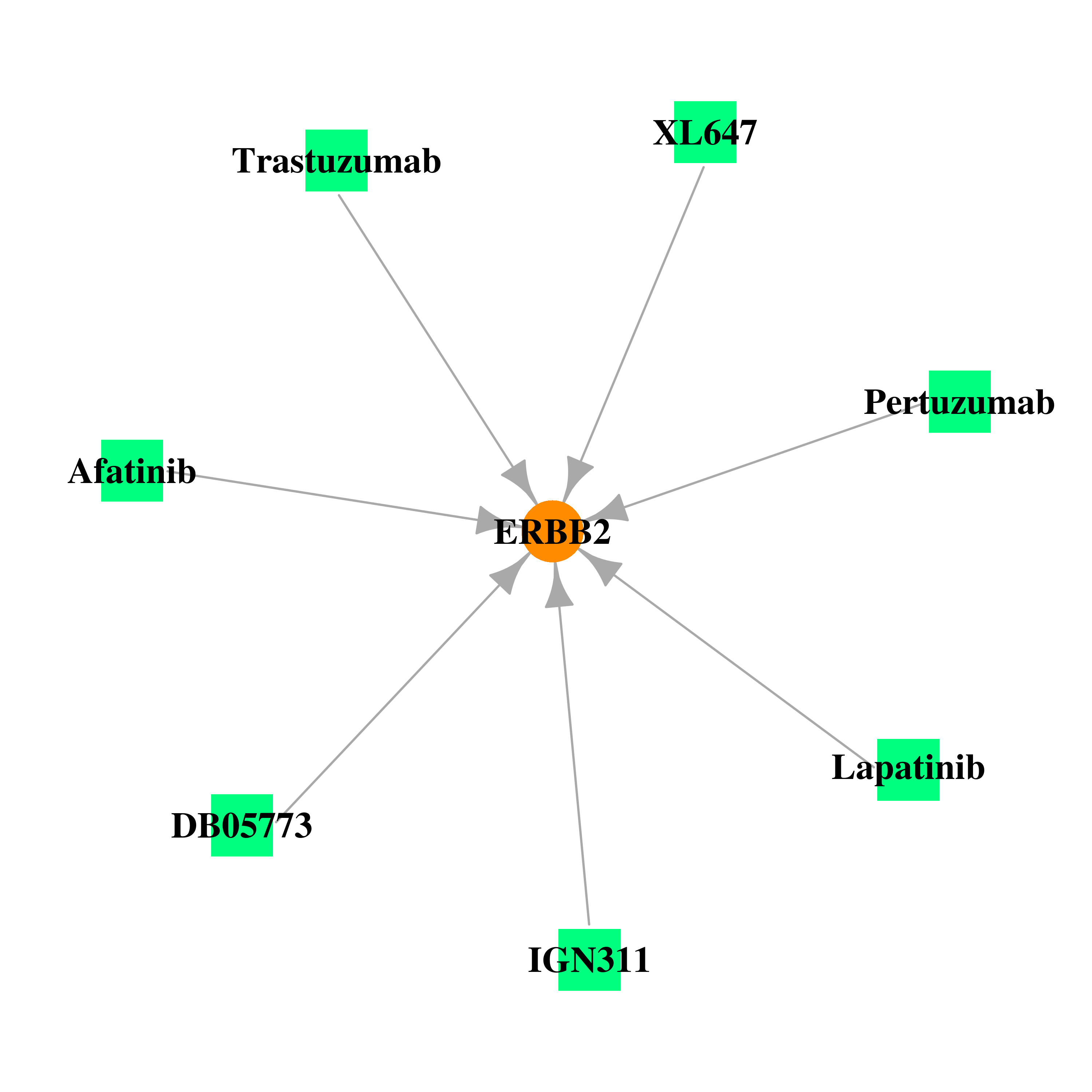
 Drug information targeting mutLBSgene (Approved drugs only)
Drug information targeting mutLBSgene (Approved drugs only)




 Gene-centered ligand-gene interaction network
Gene-centered ligand-gene interaction network 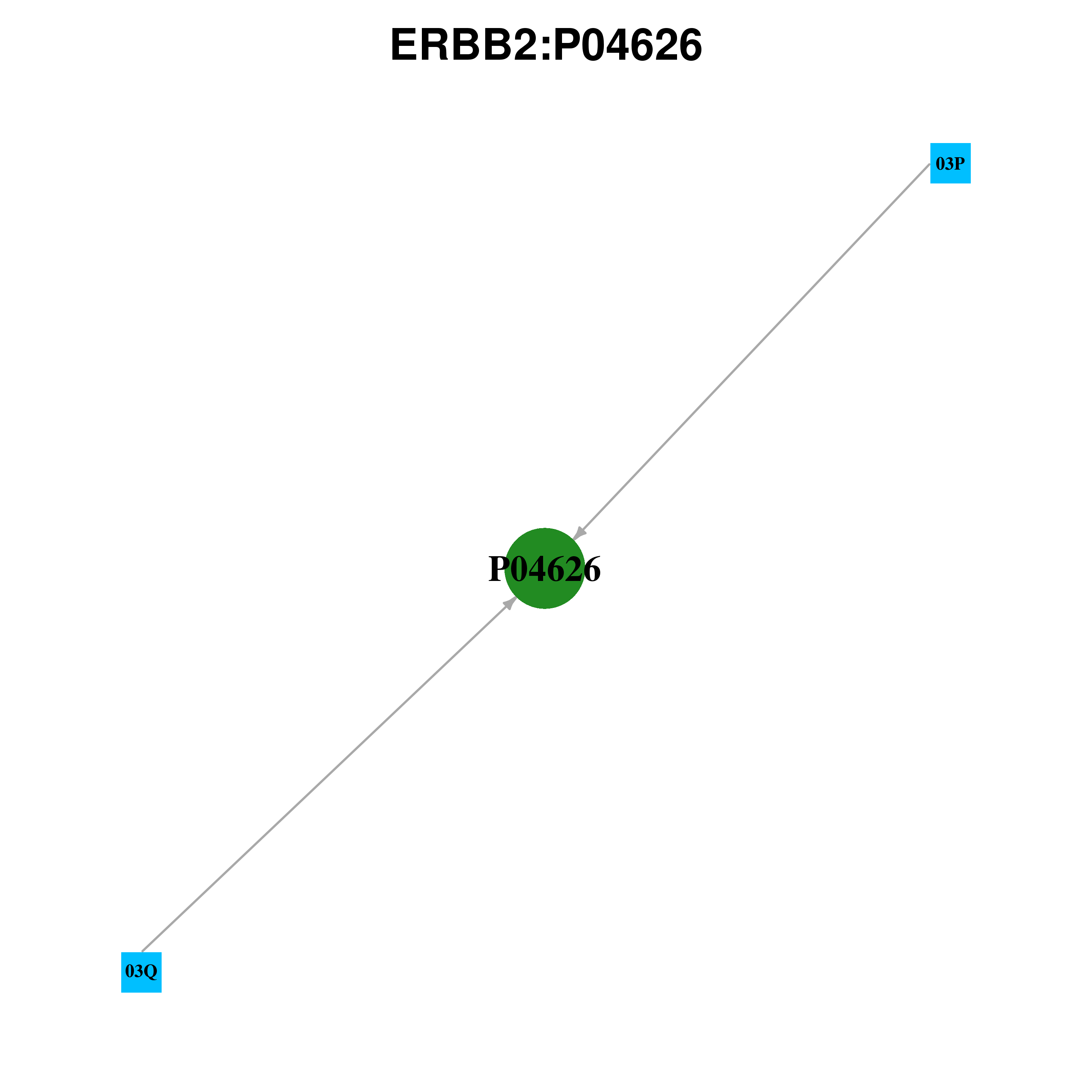
 Ligands binding to mutated ligand binding site of ERBB2 go to BioLip
Ligands binding to mutated ligand binding site of ERBB2 go to BioLip Multiple alignments for P04626 in multiple species
Multiple alignments for P04626 in multiple species 
