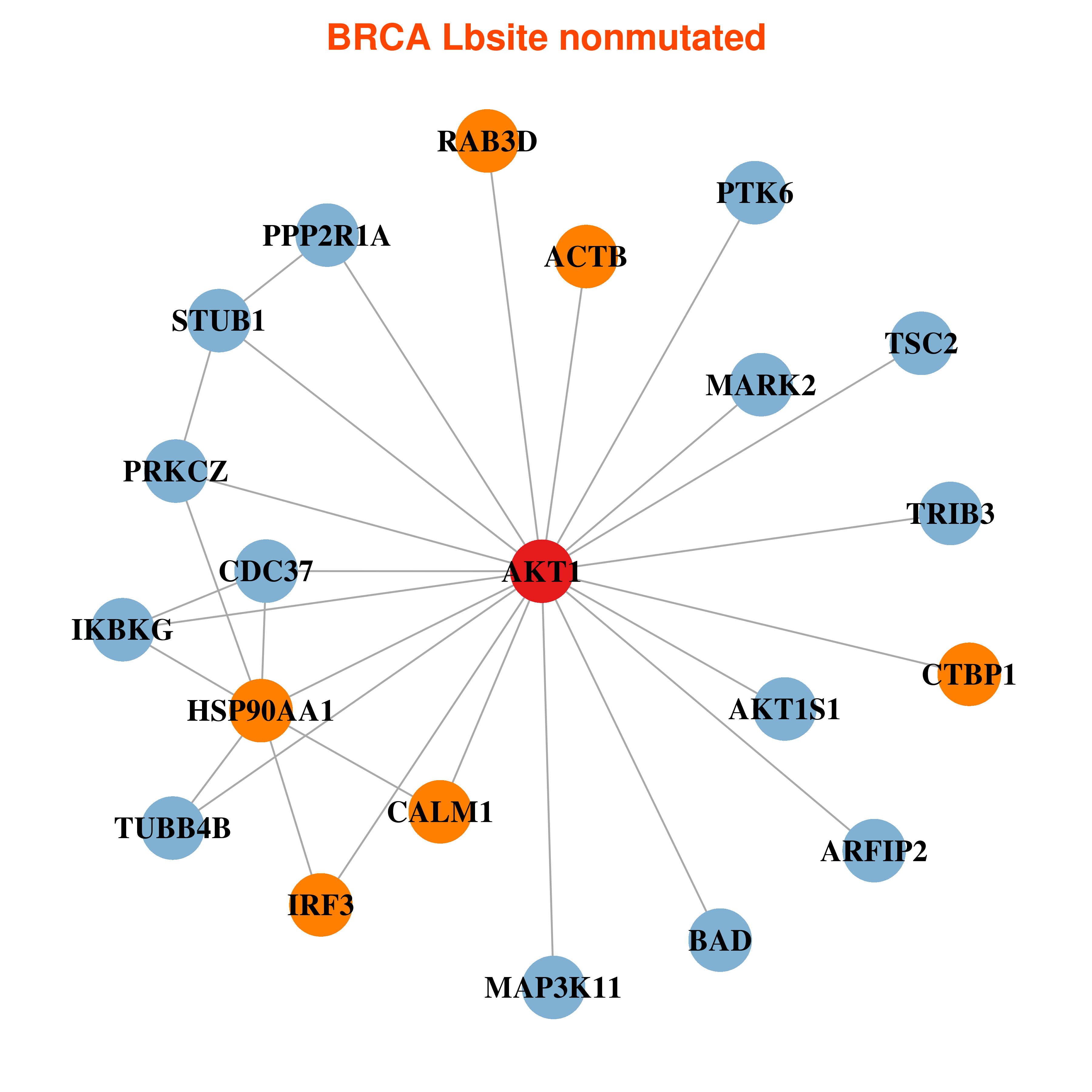
|
mutLBSgeneDB |
| |
| |
| |
| |
| |
| |
|
| Gene summary for AKT1 |
 Gene summary Gene summary |
| Basic gene Info. | Gene symbol | AKT1 |
| Gene name | v-akt murine thymoma viral oncogene homolog 1 | |
| Synonyms | AKT|CWS6|PKB|PKB-ALPHA|PRKBA|RAC|RAC-ALPHA | |
| Cytomap | UCSC genome browser: 14q32.32 | |
| Type of gene | protein-coding | |
| RefGenes | NM_001014431.1, NM_001014432.1,NM_005163.2, | |
| Description | AKT1mPKB alphaRAC-PK-alphaRAC-alpha serine/threonine-protein kinaseprotein kinase B alphaproto-oncogene c-Aktrac protein kinase alpha | |
| Modification date | 20141222 | |
| dbXrefs | MIM : 164730 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000142208 | ||
| HPRD : 01261 | ||
| Vega : OTTHUMG00000170795 | ||
| Protein | UniProt: P31749 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_AKT1 | |
| BioGPS: 207 | ||
| Pathway | NCI Pathway Interaction Database: AKT1 | |
| KEGG: AKT1 | ||
| REACTOME: AKT1 | ||
| Pathway Commons: AKT1 | ||
| Context | iHOP: AKT1 | |
| ligand binding site mutation search in PubMed: AKT1 | ||
| UCL Cancer Institute: AKT1 | ||
| Assigned class in mutLBSgeneDB | A: This gene has a literature evidence and it belongs to targetable_mutLBSgenes. | |
| References showing study about ligand binding site mutation for AKT1. | 1. "Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007 Jul 26;448(7152):439-44. Epub 2007 Jul 4. PubMed PMID: 17611497." 17611497 2. "Deyle KM, Farrow B, Qiao Hee Y, Work J, Wong M, Lai B, Umeda A, Millward SW, Nag A, Das S, Heath JR. A protein-targeting strategy used to develop a selective inhibitor of the E17K point mutation in the PH domain of Akt1. Nat Chem. 2015 May;7(5):455-62. doi: 10.1038/nchem.2223. Epub 2015 Apr 13. PubMed PMID: 25901825; PubMed Central PMCID: PMC4408887." 25901825 | |
 Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez |
| GO ID | GO Term | PubMed ID | GO:0001934 | positive regulation of protein phosphorylation | 19057511 | GO:0006468 | protein phosphorylation | 11994271 | GO:0016310 | phosphorylation | 20333297 | GO:0018105 | peptidyl-serine phosphorylation | 16139227 | GO:0030307 | positive regulation of cell growth | 19203586 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle | 18483258 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 19667065 | GO:0035556 | intracellular signal transduction | 14749367 | GO:0043066 | negative regulation of apoptotic process | 19203586 | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 20011604 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity | 19057511 | GO:0070141 | response to UV-A | 18483258 |
| Top |
| Ligand binding site mutations for AKT1 |
 Lollipop-style diagram of mutations at LBS in amino-acid sequence. Lollipop-style diagram of mutations at LBS in amino-acid sequence. We represented ligand binding site mutations only. (You can see big image via clicking.) |
 |
 Cancer type specific mutLBS sorted by frequency Cancer type specific mutLBS sorted by frequency |
| LBS | AAchange of nsSNV | Cancer type | # samples | E17 | E17K | BRCA | 18 | L52 | L52R | BRCA | 1 | F55 | F55L | COAD | 1 | T211 | T211I | GBM | 1 | E17 | E17K | HNSC | 1 | L52 | L52R | KIRC | 1 | E17 | E17K | LUSC | 1 | E17 | E17K | PRAD | 1 | E17 | E17K | SKCM | 1 | E17 | E17K | STAD | 1 | Q79 | Q79K | UCEC | 1 | W80 | W80R | UCEC | 1 |
| cf) Cancer type abbreviation. BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, COAD: Colon adenocarcinoma, GBM: Glioblastoma multiforme, LGG: Brain lower grade glioma, HNSC: Head and neck squamous cell carcinoma, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute myeloid leukemia, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PRAD: Prostate adenocarcinoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, THCA: Thyroid carcinoma, UCEC: Uterine corpus endometrial carcinoma. |
 Clinical information for AKT1 from My Cancer Genome. Clinical information for AKT1 from My Cancer Genome. |
| The E17K mutation results in an amino acid change at position 17 in AKT1, from a glutamic acid (E) to a lysine (K). This mutation occurs within the pleckstrin homology domain (PHD) of AKT1 and results in activation of the phosphatidylinositol 3-kinase (PI3K) pathway. In vitro studies suggest that the AKT1 E17K mutation is less sensitive than wild type AKT1 to inhibition by the experimental AKT inhibitor VIII, a non-ATP competitive agent which requires a functional pleckstrin homology domain (Carpten et al. 2007). Other AKT inhibitors, both allosteric and catalytic, are in clinical development.In preclinical experiments using a cell line harboring BRAF V600E and AKT1 E17K mutations (note that there are no co-occurring AKT1 and BRAF variants in any breast cancer sample in cBioPortal), the AKT1 E17K mutation was associated with sensitivity to GSK2141795B (a tool compound related to the AKT1 inhibitor uprosertib, GSK214179;Lassen et al. 2014).Balko, J., I. Mayer, M. Levy, C. Arteaga. 2015. AKT1 c.49G>A (E17K) Mutation in Breast Cancer. My Cancer Genome https://www.mycancergenome.org/content/disease/breast-cancer/akt1/23/ (Updated October 12). |
| Top |
| Protein structure related information for AKT1 |
 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Mupro score denotes assessment of the effect of mutations on thermodynamic stability. (ΔΔE<0: mutation decreases stability, ΔΔE>0: mutation increases stability) |
 : nsSNV at non-LBS : nsSNV at non-LBS : nsSNV at LBS : nsSNV at LBS |
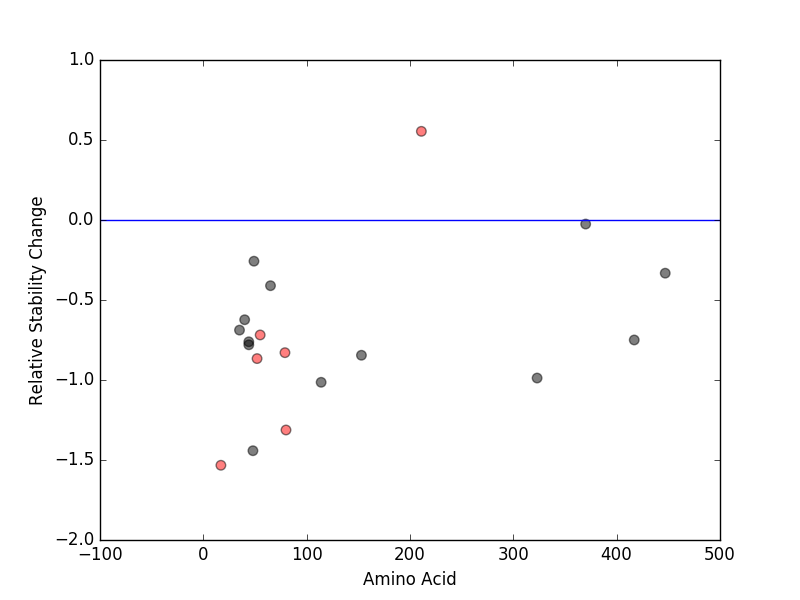 |
 nsSNVs sorted by the relative stability change of protein structure by each mutation nsSNVs sorted by the relative stability change of protein structure by each mutation Blue: mutations of positive stability change. and red : the most recurrent mutation for this gene. |
| LBS | AAchange of nsSNV | Relative stability change | T211 | T211I | 0.55415848 | E17 | E17K | -1.5329426 | W80 | W80R | -1.3124478 | L52 | L52R | -0.86616902 | Q79 | Q79K | -0.82920041 | F55 | F55L | -0.7184452 |
| (MuPro1.1: Jianlin Cheng et al., Prediction of Protein Stability Changes for Single-Site Mutations Using Support Vector Machines, PROTEINS: Structure, Function, and Bioinformatics. 2006, 62:1125-1132) |
 Structure image for AKT1 from PDB Structure image for AKT1 from PDB |
| PDB ID | PDB title | PDB structure | 2UZS | A Transforming Mutation in the Pleckstrin homology Domain of AKT1 in Cancer (AKT1-PH_E17K) | 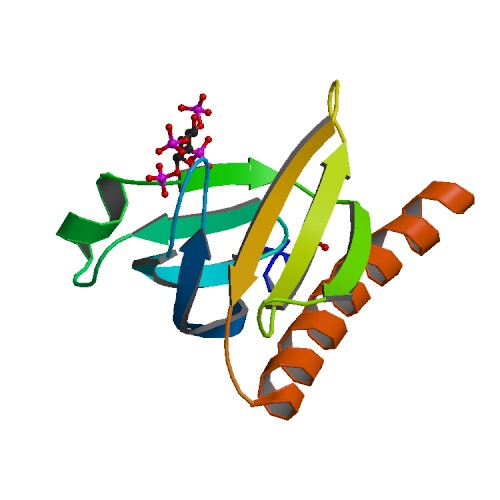 |
| Top |
| Differential gene expression and gene-gene network for AKT1 |
 Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types |
| AKT1_BRCA_DE |
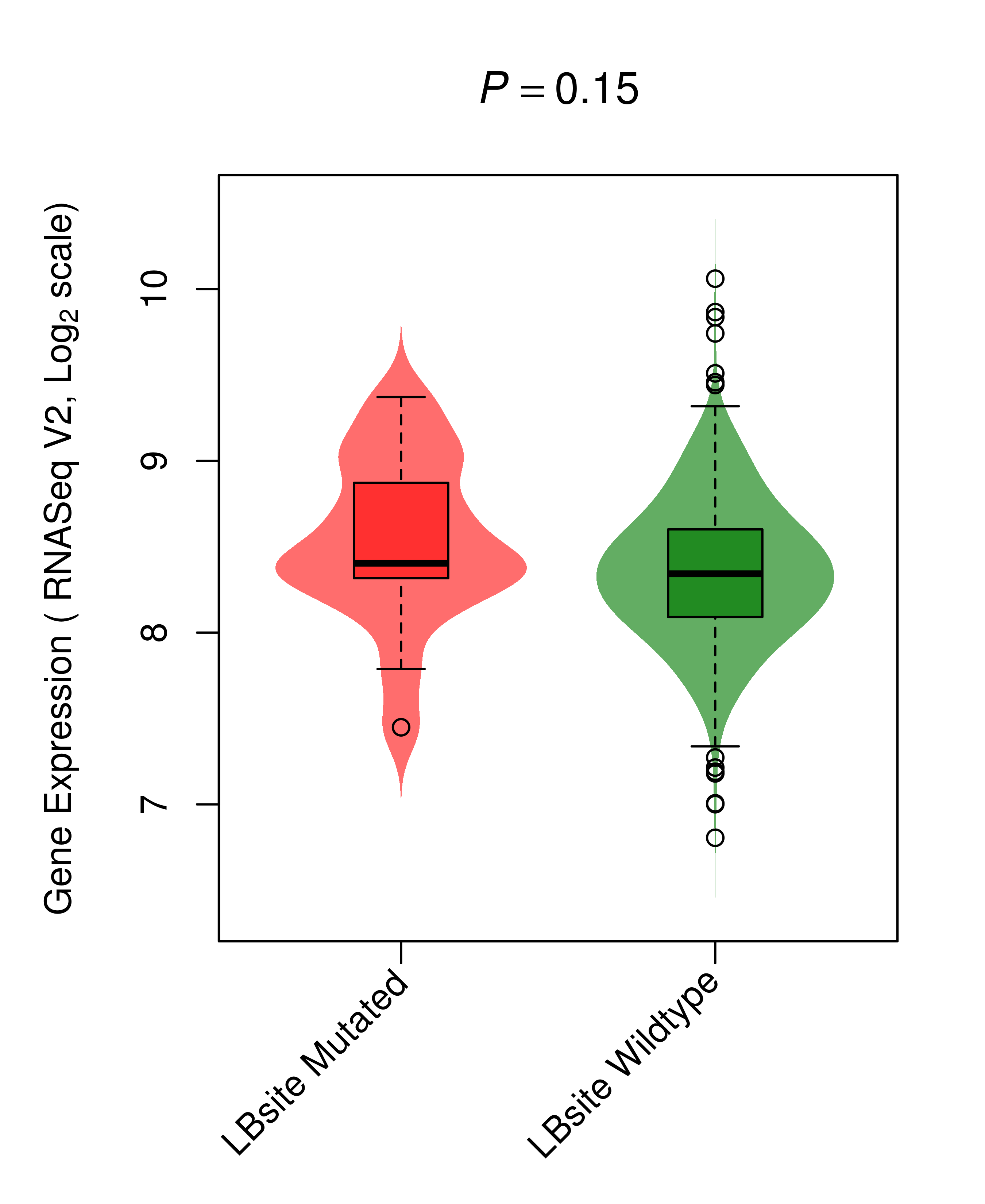 |
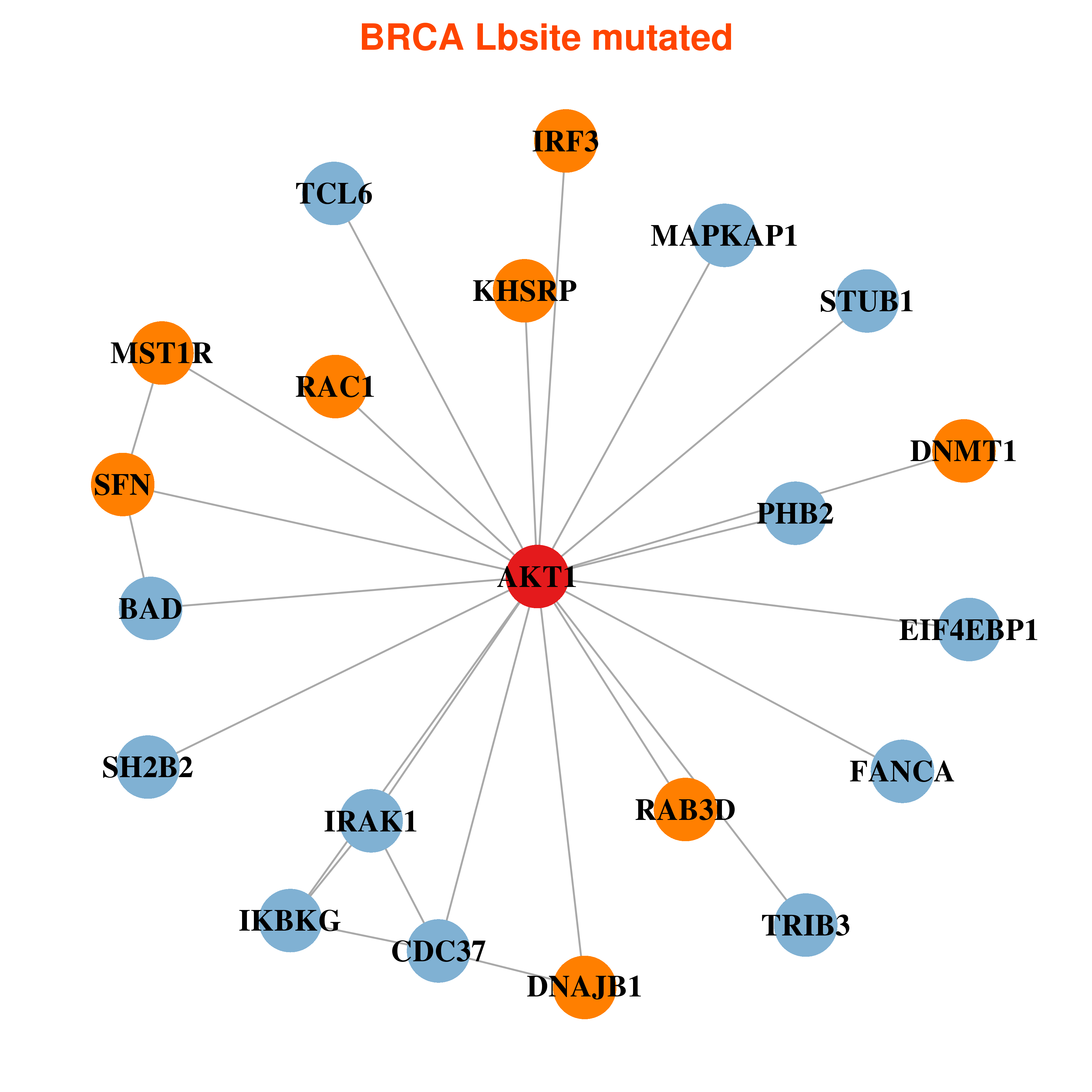
 Differential co-expressed gene network based on protein-protein interaction data (CePIN) Differential co-expressed gene network based on protein-protein interaction data (CePIN) |
| * In BRCA | |
 |
|
| Top |
| Top |
| Phenotype information for AKT1 |
 Gene level disease information (DisGeNet) Gene level disease information (DisGeNet) |
| Disease ID | Disease name | # PubMed | Association type |
| umls:C0006142 | Malignant neoplasm breast | 96 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0036341 | Schizophrenia | 47 | Biomarker, GeneticVariation |
| umls:C1458155 | Breast Neoplasms | 32 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0033578 | Prostatic Neoplasms | 29 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0024121 | Lung Neoplasms | 12 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification, Therapeutic |
| umls:C0007137 | Carcinoma, Squamous Cell | 11 | AlteredExpression, Biomarker |
| umls:C0919267 | Ovarian Neoplasms | 6 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0028754 | Obesity | 6 | Biomarker, GeneticVariation |
| umls:C0009404 | Colorectal Neoplasms | 4 | AlteredExpression, Biomarker |
| umls:C0024809 | Marijuana Abuse | 3 | Biomarker, GeneticVariation |
| umls:C0085261 | Proteus Syndrome | 2 | GeneticVariation |
| umls:C0236733 | Amphetamine-Related Disorders | 2 | Biomarker, GeneticVariation |
| umls:C0037286 | Skin Neoplasms | 2 | Biomarker |
| umls:C0026846 | Muscular Atrophy | 2 | Biomarker, Therapeutic |
| umls:C0033941 | Psychoses, Substance-Induced | 1 | Biomarker, GeneticVariation |
| umls:C0025286 | Meningioma | 1 | Biomarker, GeneticVariation |
| umls:C0032580 | Adenomatous Polyposis Coli | 1 | Biomarker |
| umls:C0428791 | Aortic Valve, Calcification of | 1 | Biomarker |
| umls:C0014544 | Epilepsy | 1 | Therapeutic |
| umls:C0020507 | Hyperplasia | 1 | Biomarker |
| umls:C0023487 | Leukemia, Promyelocytic, Acute | 1 | Biomarker |
| umls:C0030193 | Pain | 1 | Biomarker |
 Mutation level pathogenic information (ClinVar annotation) Mutation level pathogenic information (ClinVar annotation) |
| Allele ID | AA change | Clinical significance | Origin | Phenotype IDs |
| 29022 | E17K | Pathogenic | Somatic | GeneReviews:NBK99495 MedGen:C0085261 OMIM:176920 Orphanet:ORPHA744 SNOMED CT:23150001 MedGen:C0699790 SNOMED CT:269533000 MedGen:C0858252 MedGen:C0919267 OMIM:167000 SNOMED CT:123843001 |
| Top |
| Pharmacological information for AKT1 |
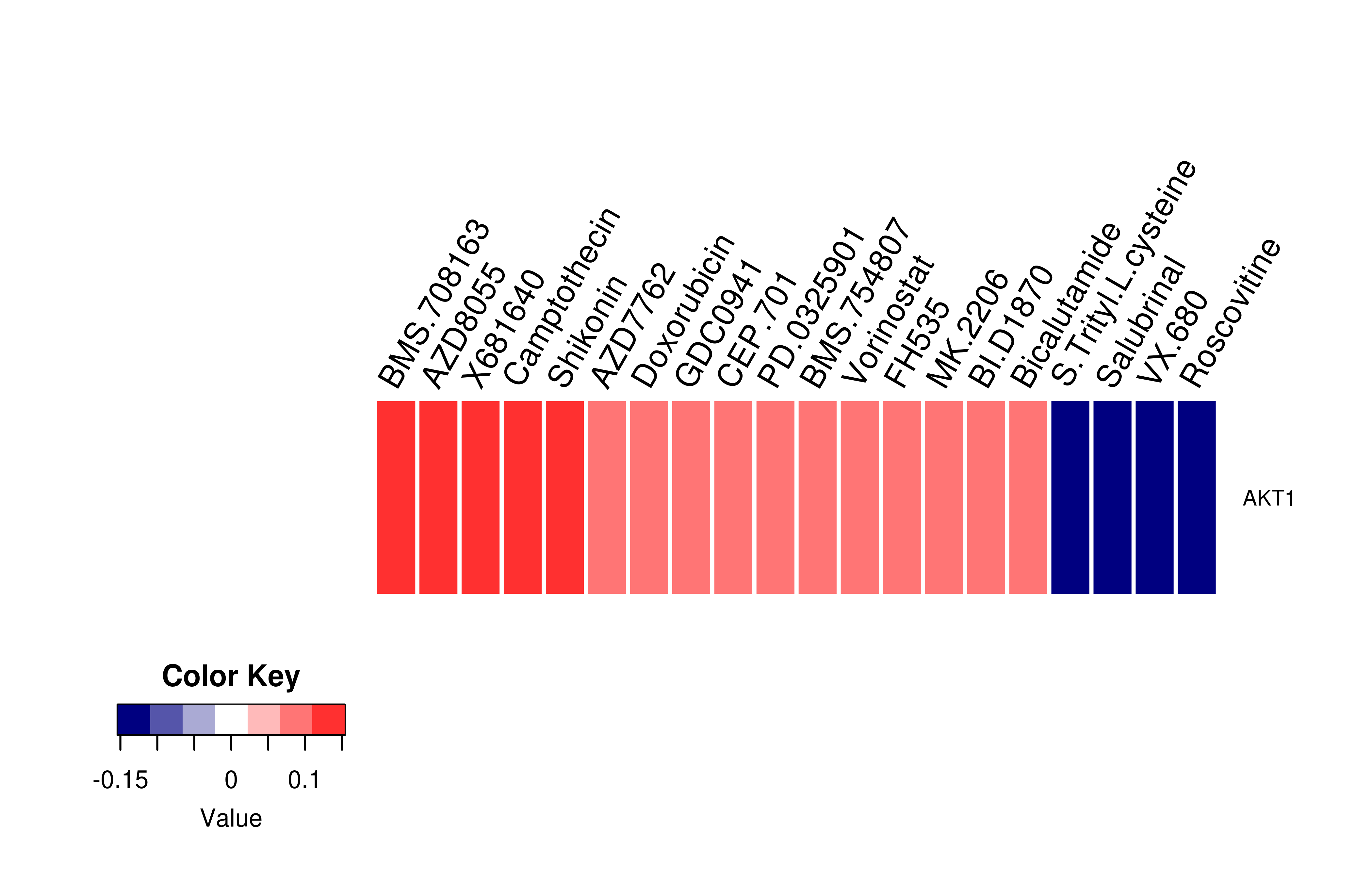
 Gene expression profile of anticancer drug treated cell-lines (CCLE) Gene expression profile of anticancer drug treated cell-lines (CCLE)Heatmap showing the correlation between gene expression and drug response across all the cell-lines. We chose the top 20 among 138 drugs.We used Pearson's correlation coefficient. |
 |
 Gene-centered drug-gene interaction network Gene-centered drug-gene interaction network |
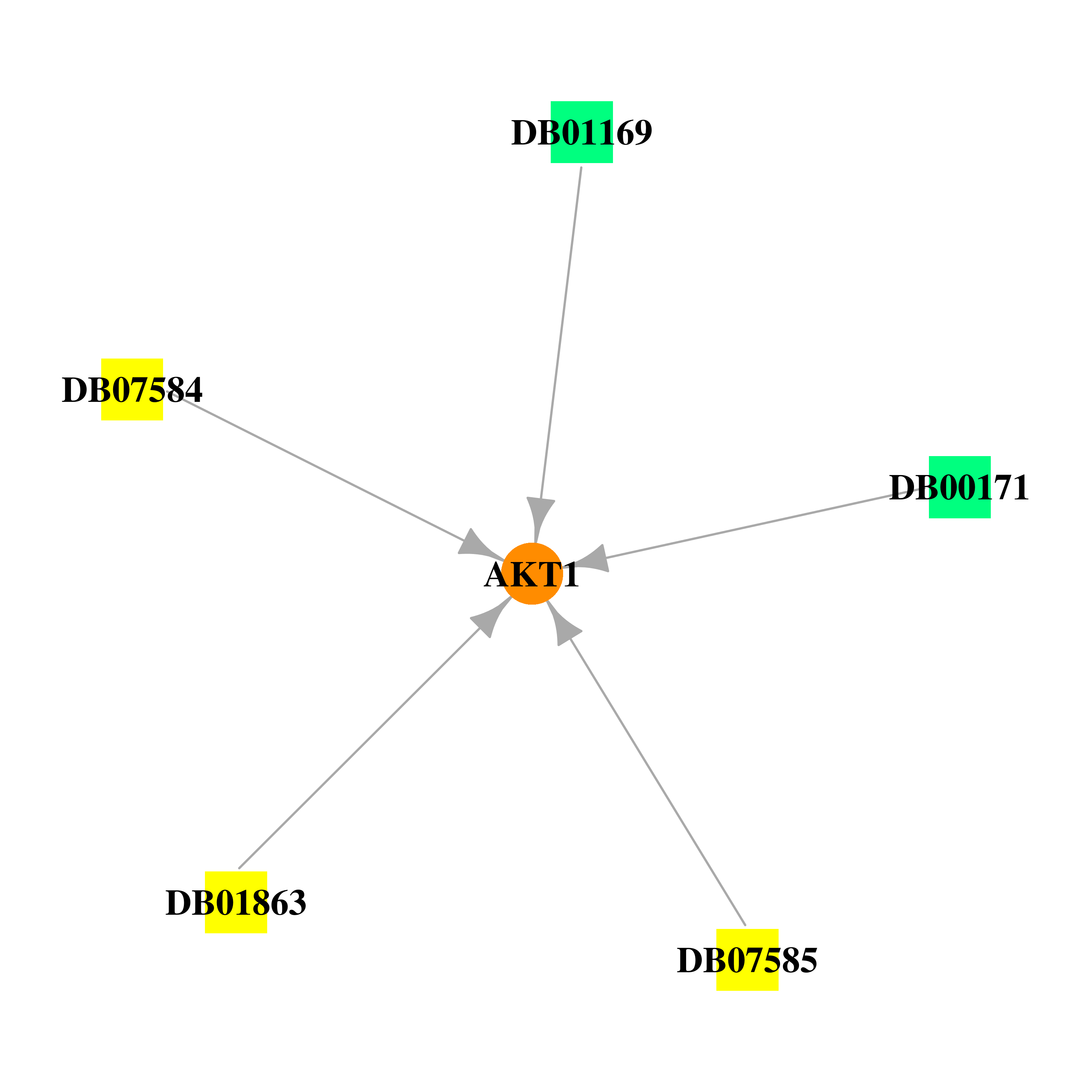 |
 Drug information targeting mutLBSgene (Approved drugs only) Drug information targeting mutLBSgene (Approved drugs only) |
| Drug status | DrugBank ID | Name | Type | Drug structure |
| Approved|nutraceutical | DB00171 | Adenosine triphosphate | Small molecule | 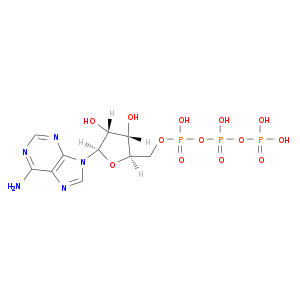 |
| Approved|investigational | DB01169 | Arsenic trioxide | Small molecule |  |
| Experimental | DB01863 | Inositol 1,3,4,5-Tetrakisphosphate | Small molecule |  |
| Experimental | DB07584 | N-[2-(5-methyl-4H-1,2,4-triazol-3-yl)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine | Small molecule |  |
| Experimental | DB07585 | 5-(5-chloro-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine | Small molecule |  |
 Gene-centered ligand-gene interaction network Gene-centered ligand-gene interaction network |
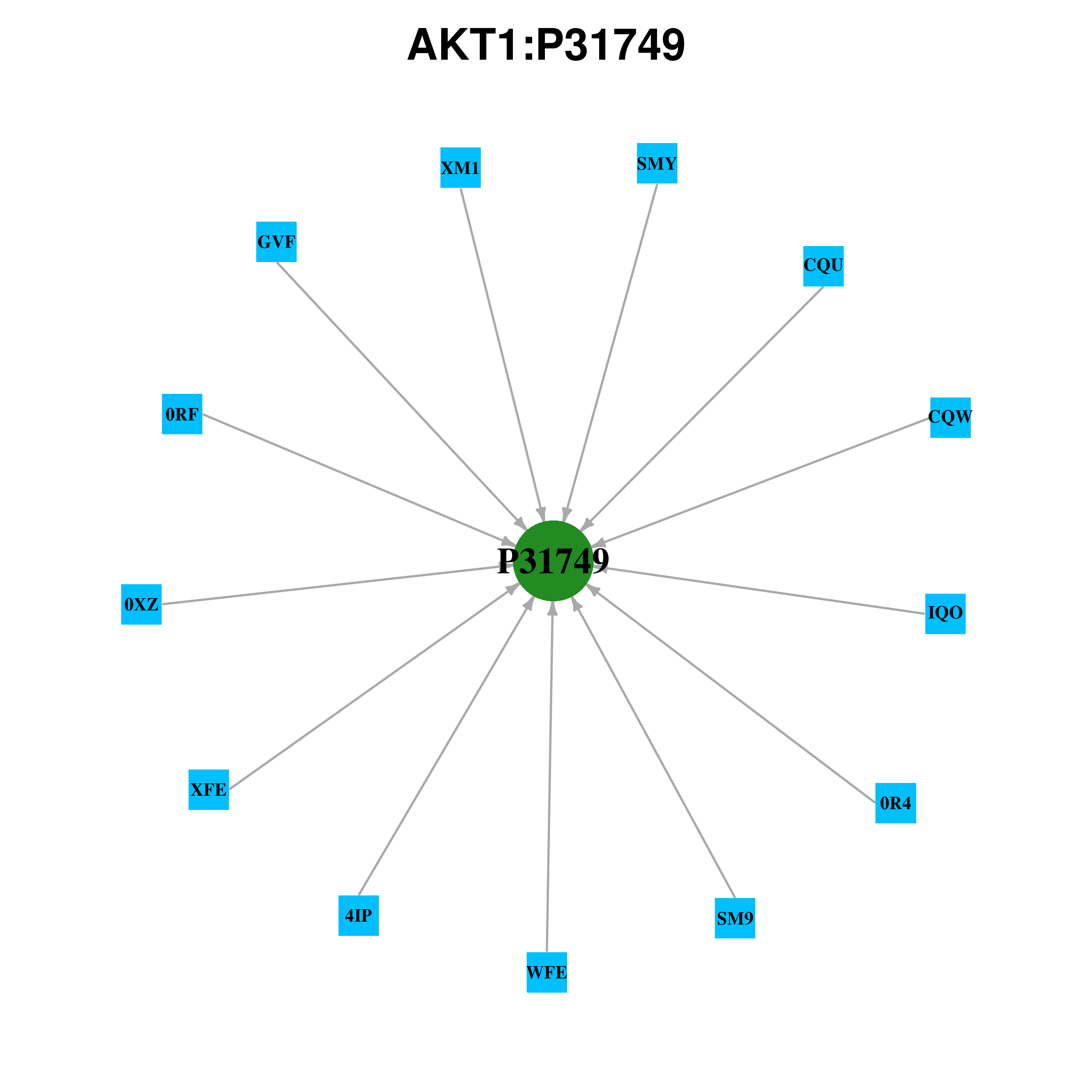 |
 Ligands binding to mutated ligand binding site of AKT1 go to BioLip Ligands binding to mutated ligand binding site of AKT1 go to BioLip |
| Ligand ID | Ligand short name | Ligand long name | PDB ID | PDB name | mutLBS | GVF | BENZENE-1,2,3,4-TETRAYL TETRAKIS[DIHYDROGEN(PHOSPHATE)] | 2uvm | A | E17 F55 | 4IP | INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | 1h10 | A | E17 L52 F55 | 4IP | INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | 1unq | A | E17 L52 F55 | 0R4 | N-(4-{5-[3-(ACETYLAMINO)PHENYL]-2-(2-AMINOPYRIDIN-3- YL)-3H-IMIDAZO[4,5-B]PYRIDIN-3-YL}BENZYL)-3- FLUOROBENZAMIDE | 4ejn | A | Q79 W80 T211 | CQU | N-[2-(5-METHYL-4H-1,2,4-TRIAZOL-3-YL)PHENYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE | 3cqu | A | T211 | CQW | 5-(5-CHLORO-7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)-4,5,6,7-TETRAHYDRO-1H-IMIDAZO[4,5-C]PYRIDINE | 3cqw | A | T211 | XFE | (3R)-1-(5-METHYL-7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)PYRROLIDIN-3-AMINE | 3mv5 | A | T211 | WFE | N-{[(3S)-3-AMINO-1-(5-ETHYL-7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)PYRROLIDIN-3-YL]METHYL}-2,4-DIFLUOROBENZAMIDE | 3mvh | A | T211 | XM1 | (2S)-2-(4-CHLOROBENZYL)-3-OXO-3-[4-(7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)PIPERAZIN-1-YL]PROPAN-1-AMINE | 3ocb | A | T211 | SMY | (2R)-3-(1H-INDOL-3-YL)-1-{4-[(5S)-5-METHYL-5,7-DIHYDROTHIENO[3,4-D]PYRIMIDIN-4-YL]PIPERAZIN-1-YL}-1-OXOPROPAN-2-AMINE | 3ow4 | A | T211 | SMY | (2R)-3-(1H-INDOL-3-YL)-1-{4-[(5S)-5-METHYL-5,7-DIHYDROTHIENO[3,4-D]PYRIMIDIN-4-YL]PIPERAZIN-1-YL}-1-OXOPROPAN-2-AMINE | 3ow4 | B | T211 | SM9 | N-(2-ETHOXYETHYL)-N-{(2S)-2-HYDROXY-3-[(5R)-2-(QUINAZOLIN-4-YL)-2,7-DIAZASPIRO[4.5]DEC-7-YL]PROPYL}-2,6-DIMETHYLBENZENESULFONAMIDE | 3qkm | A | T211 | 0RF | (2S)-2-(4-CHLOROPHENYL)-1-{4-[(5R,7R)-7-HYDROXY-5- METHYL-6,7-DIHYDRO-5H-CYCLOPENTA[D]PYRIMIDIN-4- YL]PIPERAZIN-1-YL}-3-(PROPAN-2-YLAMINO)PROPAN-1-ONE | 4ekl | A | T211 | 0XZ | 4-AMINO-N-[(1S)-1-(4-CHLOROPHENYL)-3-HYDROXYPROPYL]-1- (7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)PIPERIDINE-4- CARBOXAMIDE | 4gv1 | A | T211 | IQO | 1-{1-[4-(6-PHENYL-1H-IMIDAZO[4,5-G]QUINOXALIN-7-YL)BENZYL]PIPERIDIN-4-YL}-1,3-DIHYDRO-2H-BENZIMIDAZOL-2-ONE | 3o96 | A | W80 T211 |
| Top |
| Conservation information for LBS of AKT1 |
 Multiple alignments for P31749 in multiple species Multiple alignments for P31749 in multiple species |
| LBS | AA sequence | # species | Species | A177 | TGRYYAMKILK | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | A177 | TQKLYAIKILK | 1 | Caenorhabditis elegans | A177 | SDKLYAIKIIR | 1 | Caenorhabditis elegans | A230 | FVMEYANGGEL | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | A230 | FVMQFANGGEL | 1 | Caenorhabditis elegans | A230 | FVMEFANGGEL | 1 | Caenorhabditis elegans | C310 | TMKTFCGTPEY | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | C310 | KTSTFCGTPEY | 2 | Caenorhabditis elegans, Caenorhabditis elegans | D274 | NVVYRDLKLEN | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | D274 | EVVYRDLKLEN | 1 | Bos taurus | D274 | DIVYRDMKLEN | 1 | Caenorhabditis elegans | D274 | NIVYRDMKLEN | 1 | Caenorhabditis elegans | D292 | HIKITDFGLCK | 5 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Caenorhabditis elegans | D292 | HIKIADFGLCK | 1 | Caenorhabditis elegans | D439 | DTRYFDEEFTA | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | D439 | DTSYFDNEFTS | 1 | Caenorhabditis elegans | D439 | DTSFFDREFTS | 1 | Caenorhabditis elegans | E17 | LHKRGEYIKTW | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | E17 | LHKKGEHIRNW | 2 | Caenorhabditis elegans, Caenorhabditis elegans | E191 | IVAKDEVAHTL | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | E191 | IIAREEVAHTL | 1 | Caenorhabditis elegans | E191 | VVDRSEVAHTL | 1 | Caenorhabditis elegans | E228 | LCFVMEYANGG | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | E228 | LCFVMQFANGG | 1 | Caenorhabditis elegans | E228 | ICFVMEFANGG | 1 | Caenorhabditis elegans | E234 | YANGGELFFHL | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | E234 | FANGGELFTHV | 1 | Caenorhabditis elegans | E234 | FANGGELFTHL | 1 | Caenorhabditis elegans | E278 | RDLKLENLMLD | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | E278 | RDMKLENLLLD | 2 | Caenorhabditis elegans, Caenorhabditis elegans | E314 | FCGTPEYLAPE | 6 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Caenorhabditis elegans, Caenorhabditis elegans | E341 | GVVMYEMMCGR | 6 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Caenorhabditis elegans, Caenorhabditis elegans | F161 | LGKGTFGKVIL | 5 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Caenorhabditis elegans | F161 | LGQGTFGKVIL | 1 | Caenorhabditis elegans | F236 | NGGELFFHLSR | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | F236 | NGGELFTHVRK | 1 | Caenorhabditis elegans | F236 | NGGELFTHLQR | 1 | Caenorhabditis elegans | F309 | ATMKTFCGTPE | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | F309 | DKTSTFCGTPE | 2 | Caenorhabditis elegans, Caenorhabditis elegans | F438 | TDTRYFDEEFT | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | F438 | TDTSYFDNEFT | 1 | Caenorhabditis elegans | F438 | TDTSFFDREFT | 1 | Caenorhabditis elegans | F55 | SPLNNFSVAQC | 3 | Bos taurus, Mus musculus, Rattus norvegicus | F55 | APLNNFSVAQC | 1 | Homo sapiens | F55 | EPLNDFMIKDA | 1 | Caenorhabditis elegans | F55 | EPLNNFMIRDA | 1 | Caenorhabditis elegans | G157 | YLKLLGKGTFG | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | G157 | FLKVLGKGTFG | 1 | Caenorhabditis elegans | G157 | FLKVLGQGTFG | 1 | Caenorhabditis elegans | G159 | KLLGKGTFGKV | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | G159 | KVLGKGTFGKV | 1 | Caenorhabditis elegans | G159 | KVLGQGTFGKV | 1 | Caenorhabditis elegans | G16 | WLHKRGEYIKT | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | G16 | WLHKKGEHIRN | 2 | Caenorhabditis elegans, Caenorhabditis elegans | G162 | GKGTFGKVILV | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | G162 | GKGTFGKVILC | 1 | Caenorhabditis elegans | G162 | GQGTFGKVILC | 1 | Caenorhabditis elegans | G311 | MKTFCGTPEYL | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | G311 | TSTFCGTPEYL | 2 | Caenorhabditis elegans, Caenorhabditis elegans | H194 | KDEVAHTLTEN | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | H194 | REEVAHTLTEN | 1 | Caenorhabditis elegans | H194 | RSEVAHTLTEN | 1 | Caenorhabditis elegans | H354 | FYNQDHEKLFE | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | H354 | FYSKDHNKLFE | 1 | Caenorhabditis elegans | H354 | FSAKENGKLFE | 1 | Caenorhabditis elegans | I186 | LKKEVIVAKDE | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | I186 | LKKDVIIAREE | 1 | Caenorhabditis elegans | I186 | IRKEMVVDRSE | 1 | Caenorhabditis elegans | I19 | KRGEYIKTWRP | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | I19 | KKGEHIRNWRP | 2 | Caenorhabditis elegans, Caenorhabditis elegans | I290 | DGHIKITDFGL | 5 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Caenorhabditis elegans | I290 | DGHIKIADFGL | 1 | Caenorhabditis elegans | I84 | QWTTVIERTFH | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | I84 | QWTTVIERTFY | 2 | Caenorhabditis elegans, Caenorhabditis elegans | K14 | EGWLHKRGEYI | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | K14 | EGWLHKKGEHI | 1 | Caenorhabditis elegans | K14 | ESWLHKKGEHI | 1 | Caenorhabditis elegans | K158 | LKLLGKGTFGK | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | K158 | LKVLGKGTFGK | 1 | Caenorhabditis elegans | K158 | LKVLGQGTFGK | 1 | Caenorhabditis elegans | K163 | KGTFGKVILVK | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | K163 | KGTFGKVILCK | 1 | Caenorhabditis elegans | K163 | QGTFGKVILCR | 1 | Caenorhabditis elegans | K179 | RYYAMKILKKE | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | K179 | KLYAIKILKKD | 1 | Caenorhabditis elegans | K179 | KLYAIKIIRKE | 1 | Caenorhabditis elegans | K268 | YLHSEKNVVYR | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | K268 | YLHSEKEVVYR | 1 | Bos taurus | K268 | YLH-RCDIVYR | 1 | Caenorhabditis elegans | K268 | YLH-HRNIVYR | 1 | Caenorhabditis elegans | K276 | VYRDLKLENLM | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | K276 | VYRDMKLENLL | 2 | Caenorhabditis elegans, Caenorhabditis elegans | L156 | EYLKLLGKGTF | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | L156 | DFLKVLGKGTF | 1 | Caenorhabditis elegans | L156 | DFLKVLGQGTF | 1 | Caenorhabditis elegans | L181 | YAMKILKKEVI | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | L181 | YAIKILKKDVI | 1 | Caenorhabditis elegans | L181 | YAIKIIRKEMV | 1 | Caenorhabditis elegans | L210 | SRHPFLTALKY | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | L210 | CKHPFLTELKY | 1 | Caenorhabditis elegans | L210 | CVHPFLTLLKY | 1 | Caenorhabditis elegans | L264 | SALDYLHSEKN | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | L264 | SALDYLHSEKE | 1 | Bos taurus | L264 | LALGYLH-RCD | 1 | Caenorhabditis elegans | L264 | LALGYLH-HRN | 1 | Caenorhabditis elegans | L277 | YRDLKLENLML | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | L277 | YRDMKLENLLL | 2 | Caenorhabditis elegans, Caenorhabditis elegans | L295 | ITDFGLCKEGI | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | L295 | IADFGLCKEEI | 1 | Caenorhabditis elegans | L295 | ITDFGLCKEEI | 1 | Caenorhabditis elegans | L316 | GTPEYLAPEVL | 5 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Caenorhabditis elegans | L316 | GTPEYLAPEVI | 1 | Caenorhabditis elegans | L347 | MMCGRLPFYNQ | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | L347 | MMCGRLPFYSK | 1 | Caenorhabditis elegans | L347 | MMCGRLPFSAK | 1 | Caenorhabditis elegans | L52 | QRESPLNNFSV | 3 | Bos taurus, Mus musculus, Rattus norvegicus | L52 | QREAPLNNFSV | 1 | Homo sapiens | L52 | PFPEPLNDFMI | 1 | Caenorhabditis elegans | L52 | PLPEPLNNFMI | 1 | Caenorhabditis elegans | M147 | KHRVTMNEFEY | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | M147 | KHRVTMNDFEY | 1 | Bos taurus | M147 | RDKITMEDFDF | 1 | Caenorhabditis elegans | M147 | KNTVTMDDFDF | 1 | Caenorhabditis elegans | M227 | RLCFVMEYANG | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | M227 | YLCFVMQFANG | 1 | Caenorhabditis elegans | M227 | HICFVMEFANG | 1 | Caenorhabditis elegans | M281 | KLENLMLDKDG | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | M281 | KLENLLLDKDG | 1 | Caenorhabditis elegans | M281 | KLENLLLDRDG | 1 | Caenorhabditis elegans | N279 | DLKLENLMLDK | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | N279 | DMKLENLLLDK | 1 | Caenorhabditis elegans | N279 | DMKLENLLLDR | 1 | Caenorhabditis elegans | N53 | RESPLNNFSVA | 3 | Bos taurus, Mus musculus, Rattus norvegicus | N53 | REAPLNNFSVA | 1 | Homo sapiens | N53 | FPEPLNDFMIK | 1 | Caenorhabditis elegans | N53 | LPEPLNNFMIR | 1 | Caenorhabditis elegans | N54 | ESPLNNFSVAQ | 3 | Bos taurus, Mus musculus, Rattus norvegicus | N54 | EAPLNNFSVAQ | 1 | Homo sapiens | N54 | PEPLNDFMIKD | 1 | Caenorhabditis elegans | N54 | PEPLNNFMIRD | 1 | Caenorhabditis elegans | P313 | TFCGTPEYLAP | 6 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Caenorhabditis elegans, Caenorhabditis elegans | Q59 | NFSVAQCQLMK | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | Q59 | DFMIKDAATML | 1 | Caenorhabditis elegans | Q59 | NFMIRDAATVC | 1 | Caenorhabditis elegans | Q79 | IIRCLQWTTVI | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | Q79 | MVRCLQWTTVI | 1 | Caenorhabditis elegans | Q79 | IVRCLQWTTVI | 1 | Caenorhabditis elegans | R23 | YIKTWRPRYFL | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | R23 | HIRNWRPRYFM | 1 | Caenorhabditis elegans | R23 | HIRNWRPRYFI | 1 | Caenorhabditis elegans | R25 | KTWRPRYFLLK | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | R25 | RNWRPRYFMIF | 1 | Caenorhabditis elegans | R25 | RNWRPRYFILF | 1 | Caenorhabditis elegans | R273 | KNVVYRDLKLE | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | R273 | KEVVYRDLKLE | 1 | Bos taurus | R273 | CDIVYRDMKLE | 1 | Caenorhabditis elegans | R273 | RNIVYRDMKLE | 1 | Caenorhabditis elegans | R86 | TTVIERTFHVE | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | R86 | TTVIERTFYAE | 1 | Caenorhabditis elegans | R86 | TTVIERTFYAD | 1 | Caenorhabditis elegans | S205 | RVLQNSRHPFL | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | S205 | RVLQRCKHPFL | 1 | Caenorhabditis elegans | S205 | RVLYACVHPFL | 1 | Caenorhabditis elegans | T195 | DEVAHTLTENR | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | T195 | EEVAHTLTENR | 1 | Caenorhabditis elegans | T195 | SEVAHTLTENR | 1 | Caenorhabditis elegans | T211 | RHPFLTALKYS | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | T211 | KHPFLTELKYS | 1 | Caenorhabditis elegans | T211 | VHPFLTLLKYS | 1 | Caenorhabditis elegans | T291 | GHIKITDFGLC | 5 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Caenorhabditis elegans | T291 | GHIKIADFGLC | 1 | Caenorhabditis elegans | T308 | GATMKTFCGTP | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | T308 | GDKTSTFCGTP | 2 | Caenorhabditis elegans, Caenorhabditis elegans | T312 | KTFCGTPEYLA | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | T312 | STFCGTPEYLA | 2 | Caenorhabditis elegans, Caenorhabditis elegans | V164 | GTFGKVILVKE | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | V164 | GTFGKVILCKE | 1 | Caenorhabditis elegans | V164 | GTFGKVILCRE | 1 | Caenorhabditis elegans | V270 | HSEKNVVYRDL | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | V270 | HSEKEVVYRDL | 1 | Bos taurus | V270 | H-RCDIVYRDM | 1 | Caenorhabditis elegans | V270 | H-HRNIVYRDM | 1 | Caenorhabditis elegans | V271 | SEKNVVYRDLK | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | V271 | SEKEVVYRDLK | 1 | Bos taurus | V271 | -RCDIVYRDMK | 1 | Caenorhabditis elegans | V271 | -HRNIVYRDMK | 1 | Caenorhabditis elegans | W80 | IRCLQWTTVIE | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | W80 | VRCLQWTTVIE | 2 | Caenorhabditis elegans, Caenorhabditis elegans | Y18 | HKRGEYIKTWR | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | Y18 | HKKGEHIRNWR | 2 | Caenorhabditis elegans, Caenorhabditis elegans | Y229 | CFVMEYANGGE | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | Y229 | CFVMQFANGGE | 1 | Caenorhabditis elegans | Y229 | CFVMEFANGGE | 1 | Caenorhabditis elegans | Y272 | EKNVVYRDLKL | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | Y272 | EKEVVYRDLKL | 1 | Bos taurus | Y272 | RCDIVYRDMKL | 1 | Caenorhabditis elegans | Y272 | HRNIVYRDMKL | 1 | Caenorhabditis elegans | Y315 | CGTPEYLAPEV | 6 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus, Caenorhabditis elegans, Caenorhabditis elegans | Y326 | LEDNDYGRAVD | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | Y326 | LDDHDYGRCVD | 1 | Caenorhabditis elegans | Y326 | IEDIDYDRSVD | 1 | Caenorhabditis elegans | Y350 | GRLPFYNQDHE | 4 | Homo sapiens, Bos taurus, Mus musculus, Rattus norvegicus | Y350 | GRLPFYSKDHN | 1 | Caenorhabditis elegans | Y350 | GRLPFSAKENG | 1 | Caenorhabditis elegans |
 |
Copyright © 2016-Present - The University of Texas Health Science Center at Houston |