|
mutLBSgeneDB |
| |
| |
| |
| |
| |
| |
|
| Gene summary for BRD1 |
 Gene summary Gene summary |
| Basic gene Info. | Gene symbol | BRD1 |
| Gene name | bromodomain containing 1 | |
| Synonyms | BRL|BRPF1|BRPF2 | |
| Cytomap | UCSC genome browser: 22q13.33 | |
| Type of gene | protein-coding | |
| RefGenes | NM_014577.1, | |
| Description | BR140-like proteinbromodomain and PHD finger-containing protein 2bromodomain-containing protein 1 | |
| Modification date | 20141215 | |
| dbXrefs | MIM : 604589 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000100425 | ||
| HPRD : 06848 | ||
| Vega : OTTHUMG00000150288 | ||
| Protein | UniProt: O95696 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_BRD1 | |
| BioGPS: 23774 | ||
| Pathway | NCI Pathway Interaction Database: BRD1 | |
| KEGG: BRD1 | ||
| REACTOME: BRD1 | ||
| Pathway Commons: BRD1 | ||
| Context | iHOP: BRD1 | |
| ligand binding site mutation search in PubMed: BRD1 | ||
| UCL Cancer Institute: BRD1 | ||
| Assigned class in mutLBSgeneDB | B: This gene belongs to targetable_mutLBSgenes. | |
 Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez |
| GO ID | GO Term | PubMed ID | GO:0043966 | histone H3 acetylation | 16387653 |
| Top |
| Ligand binding site mutations for BRD1 |
 Lollipop-style diagram of mutations at LBS in amino-acid sequence. Lollipop-style diagram of mutations at LBS in amino-acid sequence. We represented ligand binding site mutations only. (You can see big image via clicking.) |
 |
 Cancer type specific mutLBS sorted by frequency Cancer type specific mutLBS sorted by frequency |
| LBS | AAchange of nsSNV | Cancer type | # samples | C385 | C385Y | BLCA | 1 | S997 | R996S | COAD | 1 | H388 | T389M | HNSC | 1 | C258 | R259C | STAD | 1 | Y940 | Y940C | STAD | 1 | F648 | V647M | STAD | 1 | C258 | R259H | STAD | 1 | C385 | A383T | UCEC | 1 | Y943 | P944Q | UCEC | 1 | C638 | C638Y | UCEC | 1 | Y599 | D598Y | UCEC | 1 | F648 | V647M | UCEC | 1 |
| cf) Cancer type abbreviation. BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, COAD: Colon adenocarcinoma, GBM: Glioblastoma multiforme, LGG: Brain lower grade glioma, HNSC: Head and neck squamous cell carcinoma, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute myeloid leukemia, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PRAD: Prostate adenocarcinoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, THCA: Thyroid carcinoma, UCEC: Uterine corpus endometrial carcinoma. |
| Top |
| Protein structure related information for BRD1 |
 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Mupro score denotes assessment of the effect of mutations on thermodynamic stability. (ΔΔE<0: mutation decreases stability, ΔΔE>0: mutation increases stability) |
 : nsSNV at non-LBS : nsSNV at non-LBS : nsSNV at LBS : nsSNV at LBS |
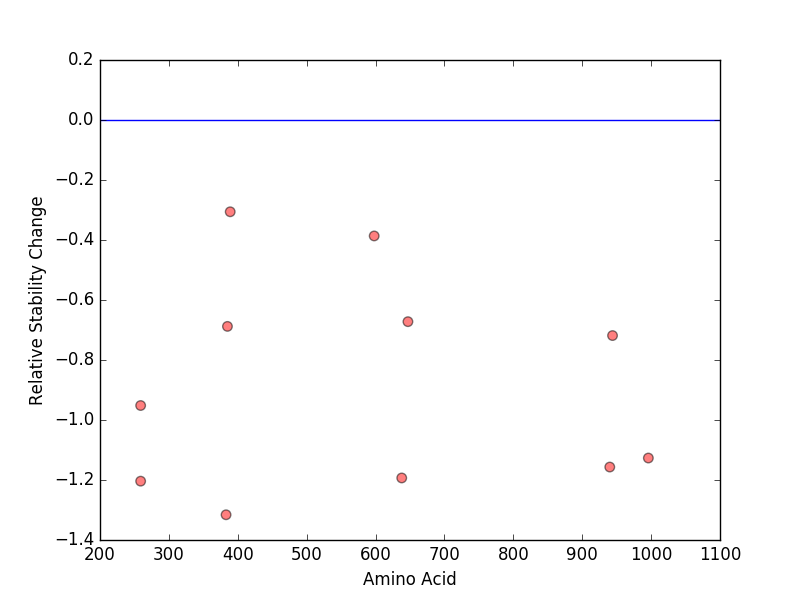 |
 nsSNVs sorted by the relative stability change of protein structure by each mutation nsSNVs sorted by the relative stability change of protein structure by each mutation Blue: mutations of positive stability change. and red : the most recurrent mutation for this gene. |
| LBS | AAchange of nsSNV | Relative stability change | C385 | A383T | -1.3158874 | C258 | R259H | -1.2038794 | C638 | C638Y | -1.1932111 | Y940 | Y940C | -1.1568137 | S997 | R996S | -1.1267772 | C258 | R259C | -0.95172878 | Y943 | P944Q | -0.71853522 | C385 | C385Y | -0.68796915 | F648 | V647M | -0.67221786 | Y599 | D598Y | -0.38647451 | H388 | T389M | -0.30594802 |
| (MuPro1.1: Jianlin Cheng et al., Prediction of Protein Stability Changes for Single-Site Mutations Using Support Vector Machines, PROTEINS: Structure, Function, and Bioinformatics. 2006, 62:1125-1132) |
 Structure image for BRD1 from PDB Structure image for BRD1 from PDB |
| Top |
| Differential gene expression and gene-gene network for BRD1 |
 Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types |
 Differential co-expressed gene network based on protein-protein interaction data (CePIN) Differential co-expressed gene network based on protein-protein interaction data (CePIN) |
| Top |
| Top |
| Phenotype information for BRD1 |
 Gene level disease information (DisGeNet) Gene level disease information (DisGeNet) |
| Disease ID | Disease name | # PubMed | Association type |
| umls:C0010606 | Carcinoma, Adenoid Cystic | 1 | Biomarker |
 Mutation level pathogenic information (ClinVar annotation) Mutation level pathogenic information (ClinVar annotation) |
| Allele ID | AA change | Clinical significance | Origin | Phenotype IDs |
| Top |
| Pharmacological information for BRD1 |
 Gene expression profile of anticancer drug treated cell-lines (CCLE) Gene expression profile of anticancer drug treated cell-lines (CCLE)Heatmap showing the correlation between gene expression and drug response across all the cell-lines. We chose the top 20 among 138 drugs.We used Pearson's correlation coefficient. |
 |
 Drug information targeting mutLBSgene (Approved drugs only) Drug information targeting mutLBSgene (Approved drugs only) |
| Drug status | DrugBank ID | Name | Type | Drug structure |
 Gene-centered ligand-gene interaction network Gene-centered ligand-gene interaction network |
 |
 Ligands binding to mutated ligand binding site of BRD1 go to BioLip Ligands binding to mutated ligand binding site of BRD1 go to BioLip |
| Ligand ID | Ligand short name | Ligand long name | PDB ID | PDB name | mutLBS | ZN | ZINC(2+) | 2lq6 | A | C385 H388 | MB3 | N-METHYLPYRROLIDIN-2-ONE | 3rcw | C | C638 F648 | OCS | L-CYSTEIC ACID | 3lyi | A | S997 | 4K8 | ISOQUINOLINE-3-CARBOXYLIC ACID | 4z02 | A | Y940 Y943 S997 | 4K8 | ISOQUINOLINE-3-CARBOXYLIC ACID | 4z02 | B | Y943 S997 |
| Top |
| Conservation information for LBS of BRD1 |
 Multiple alignments for O95696 in multiple species Multiple alignments for O95696 in multiple species |
| LBS | AA sequence | # species | Species |
 |
Copyright © 2016-Present - The University of Texas Health Science Center at Houston |
