|
mutLBSgeneDB |
| |
| |
| |
| |
| |
| |
|
| Gene summary for KRAS |
 Gene summary Gene summary |
| Basic gene Info. | Gene symbol | KRAS |
| Gene name | Kirsten rat sarcoma viral oncogene homolog | |
| Synonyms | C-K-RAS|CFC2|K-RAS2A|K-RAS2B|K-RAS4A|K-RAS4B|KI-RAS|KRAS1|KRAS2|NS|NS3|RASK2 | |
| Cytomap | UCSC genome browser: 12p12.1 | |
| Type of gene | protein-coding | |
| RefGenes | NM_004985.4, NM_033360.3, | |
| Description | GTPase KRasK-Ras 2K-ras p21 proteinPR310 c-K-ras oncogenec-Ki-rasc-Kirsten-ras proteincellular c-Ki-ras2 proto-oncogeneoncogene KRAS2transforming protein p21v-Ki-ras2 Kirsten rat sarcoma 2 viral oncogene homolog | |
| Modification date | 20141222 | |
| dbXrefs | MIM : 190070 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000133703 | ||
| HPRD : 01817 | ||
| Vega : OTTHUMG00000171193 | ||
| Protein | UniProt: P01116 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_KRAS | |
| BioGPS: 3845 | ||
| Pathway | NCI Pathway Interaction Database: KRAS | |
| KEGG: KRAS | ||
| REACTOME: KRAS | ||
| Pathway Commons: KRAS | ||
| Context | iHOP: KRAS | |
| ligand binding site mutation search in PubMed: KRAS | ||
| UCL Cancer Institute: KRAS | ||
| Assigned class in mutLBSgeneDB | A: This gene has a literature evidence and it belongs to targetable_mutLBSgenes. | |
| References showing study about ligand binding site mutation for KRAS. | 1. Maurer, T., Garrenton, L. S., Oh, A., Pitts, K., Anderson, D. J., Skelton, N. J., ... & Ludlam, M. J. (2012). Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.Proceedings of the National Academy of Sciences, 109(14), 5299-5304. 22431598 | |
 Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez |
| GO ID | GO Term | PubMed ID |
| Top |
| Ligand binding site mutations for KRAS |
 Cancer type specific mutLBS sorted by frequency Cancer type specific mutLBS sorted by frequency |
| LBS | AAchange of nsSNV | Cancer type | # samples | G12 | G12D | COAD | 31 | G12 | G12C | LUAD | 30 | G12 | G12V | COAD | 23 | G12 | G12D | UCEC | 16 | G12 | G12V | LUAD | 15 | G12 | G12V | UCEC | 12 | G13 | G13D | COAD | 10 | G13 | G13D | STAD | 9 | A146 | A146T | COAD | 8 | G12 | G12D | STAD | 7 | G13 | G13D | UCEC | 7 | G12 | G12C | COAD | 6 | K117 | K117N | COAD | 5 | G12 | G12A | LUAD | 5 | G12 | G12A | UCEC | 5 | G12 | G12V | BRCA | 3 | G12 | G12S | COAD | 3 | G12 | G12A | COAD | 3 | Q61 | Q61L | LUAD | 3 | G12 | G12D | LUAD | 3 | Q61 | Q61H | STAD | 3 | Q70,R68 | D69G | COAD | 2 | Q61 | Q61L | COAD | 2 | K5 | K5R | COAD | 2 | Q61 | Q61K | COAD | 2 | A146 | A146V | COAD | 2 | M72 | M72T | COAD | 2 | S17 | S17G | COAD | 2 | M72 | M72V | COAD | 2 | G12 | G12D | GBM | 2 | G12 | G12D | LAML | 2 | G12 | G12V | OV | 2 | A146 | A146T | STAD | 2 | G12 | G12S | STAD | 2 | Q61 | Q61H | UCEC | 2 | G12 | G12C | UCEC | 2 | G12 | G12C | BRCA | 1 | G12 | G12D | BRCA | 1 | T35 | T35A | COAD | 1 | G13 | G13C | COAD | 1 | G12 | G12R | COAD | 1 | R68 | R68S | COAD | 1 | A18 | T20A | COAD | 1 | E62 | E63K | COAD | 1 | G12 | G10R | COAD | 1 | V7 | V9F | COAD | 1 | F28 | H27N | COAD | 1 | E62 | E62G | COAD | 1 | P34 | P34L | COAD | 1 | V14 | V14A | COAD | 1 | R68 | R68G | COAD | 1 | A146 | A146G | COAD | 1 | V7 | V8A | COAD | 1 | V14 | V14G | COAD | 1 | Y71 | Y71C | COAD | 1 | V7 | V9I | COAD | 1 | K117 | K117E | COAD | 1 | Q61 | Q61H | KIRC | 1 | A59 | A59G | KIRC | 1 | G12 | G12D | KIRC | 1 | Q61 | Q61H | LAML | 1 | G12 | G12V | LAML | 1 | A59 | A59E | LAML | 1 | E37,T35 | I36M | LAML | 1 | A146 | A146T | LAML | 1 | G13 | G13D | LAML | 1 | G13 | G13C | LUAD | 1 | A146 | A146P | LUAD | 1 | G13 | G13D | LUAD | 1 | P34,Y32 | D33E | LUAD | 1 | D119,K117 | C118S | LUSC | 1 | Q61 | Q61H | LUSC | 1 | E62 | E62K | SKCM | 1 | Q61 | Q61R | SKCM | 1 | K117 | K117N | SKCM | 1 | G12 | G12D | SKCM | 1 | G12 | G12C | STAD | 1 | Q61 | Q61R | THCA | 1 | G12 | G12V | THCA | 1 | Q61 | Q61L | UCEC | 1 | G12 | G12S | UCEC | 1 | A18 | T20R | UCEC | 1 | G13 | G13C | UCEC | 1 | G13 | G13V | UCEC | 1 |
| cf) Cancer type abbreviation. BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, COAD: Colon adenocarcinoma, GBM: Glioblastoma multiforme, LGG: Brain lower grade glioma, HNSC: Head and neck squamous cell carcinoma, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute myeloid leukemia, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PRAD: Prostate adenocarcinoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, THCA: Thyroid carcinoma, UCEC: Uterine corpus endometrial carcinoma. |
 Clinical information for KRAS from My Cancer Genome. Clinical information for KRAS from My Cancer Genome. |
| The G12V mutation results in an amino acid substitution at position 12 in KRAS, from a glycine (G) to a valine (V). The role of KRAS mutations for selecting/prioritizing anti-cancer treatment, including cytotoxic chemotherapy and targeted agents, is unknown at this time.A clinical trial showed that the MEK inhibitor selumetinib increased responses to radioactive iodine in patients with radioactive iodine–refractory metastatic thyroid cancer, particularly in RAS-mutated tumors (Ho et al. 2013). In this trial, 5 of 5 patients with NRAS-mutated tumors achieved greater radioiodine incorporation into metastatic sites after selumetinib. Partial responses to therapeutic radioiodine were observed in 4 of the patients, and stable disease was observed in the 5th (Ho et al. 2013).While sorafenib has had beneficial effects for patients with metastatic MTC and DTC, it is unknown whether mutation status affects sensitivity to sorafenib (Hoftijzer et al. 2009, Kloos et al. 2009).Espinosa, A., J. Gilbert, J. Fagin. 2015. KRAS c.35G>T (G12V) Mutation in Thyroid Cancer.My Cancer Genome https://www.mycancergenome.org/content/disease/thyroid-cancer/kras/37/ (Updated February 17). |
| Top |
| Protein structure related information for KRAS |
 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Mupro score denotes assessment of the effect of mutations on thermodynamic stability. (ΔΔE<0: mutation decreases stability, ΔΔE>0: mutation increases stability) |
 : nsSNV at non-LBS : nsSNV at non-LBS : nsSNV at LBS : nsSNV at LBS |
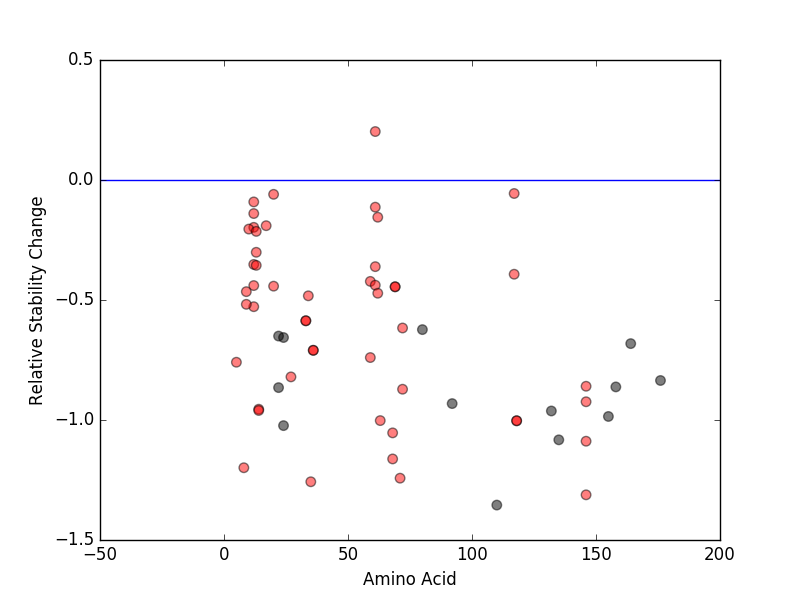 |
 nsSNVs sorted by the relative stability change of protein structure by each mutation nsSNVs sorted by the relative stability change of protein structure by each mutation Blue: mutations of positive stability change. and red : the most recurrent mutation for this gene. |
| LBS | AAchange of nsSNV | Relative stability change | Q61 | Q61L | 0.20187714 | A146 | A146G | -1.3117704 | T35 | T35A | -1.2573672 | Y71 | Y71C | -1.2424236 | V7 | V8A | -1.1989265 | R68 | R68G | -1.1622646 | A146 | A146T | -1.088413 | R68 | R68S | -1.0536319 | K117 | C118S | -1.0031905 | D119 | C118S | -1.0031905 | E62 | E63K | -1.0024674 | V14 | V14G | -0.96035751 | V14 | V14A | -0.95507363 | A146 | A146P | -0.92400051 | M72 | M72T | -0.87178184 | A146 | A146V | -0.85941844 | F28 | H27N | -0.82038307 | K5 | K5R | -0.75930727 | A59 | A59G | -0.73978146 | E37 | I36M | -0.70963745 | T35 | I36M | -0.70963745 | M72 | M72V | -0.61665747 | Y32 | D33E | -0.58652925 | P34 | D33E | -0.58652925 | G12 | G12A | -0.52787992 | V7 | V9F | -0.51801687 | P34 | P34L | -0.4825546 | E62 | E62G | -0.47181156 | V7 | V9I | -0.46520295 | R68 | D69G | -0.4449611 | Q70 | D69G | -0.4449611 | A18 | T20A | -0.4423276 | G12 | G12S | -0.4398267 | Q61 | Q61H | -0.43826644 | A59 | A59E | -0.4224792 | K117 | K117N | -0.3926214 | Q61 | Q61K | -0.36102568 | G13 | G13D | -0.3556049 | G12 | G12D | -0.35182111 | G13 | G13C | -0.30121522 | G13 | G13V | -0.21411651 | G12 | G10R | -0.20449613 | G12 | G12C | -0.19725542 | S17 | S17G | -0.19035903 | E62 | E62K | -0.15511236 | G12 | G12R | -0.13974981 | Q61 | Q61R | -0.11317789 | G12 | G12V | -0.091303772 | A18 | T20R | -0.059715487 | K117 | K117E | -0.056241973 |
| (MuPro1.1: Jianlin Cheng et al., Prediction of Protein Stability Changes for Single-Site Mutations Using Support Vector Machines, PROTEINS: Structure, Function, and Bioinformatics. 2006, 62:1125-1132) |
 Structure image for KRAS from PDB Structure image for KRAS from PDB |
| PDB ID | PDB title | PDB structure | 4TQ9 | Crystal Structure of a GDP-bound G12V Oncogenic Mutant of Human GTPase Kras | 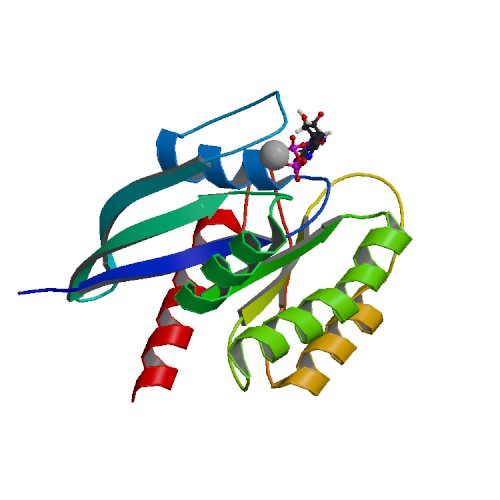 |
| Top |
| Differential gene expression and gene-gene network for KRAS |
 Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types |
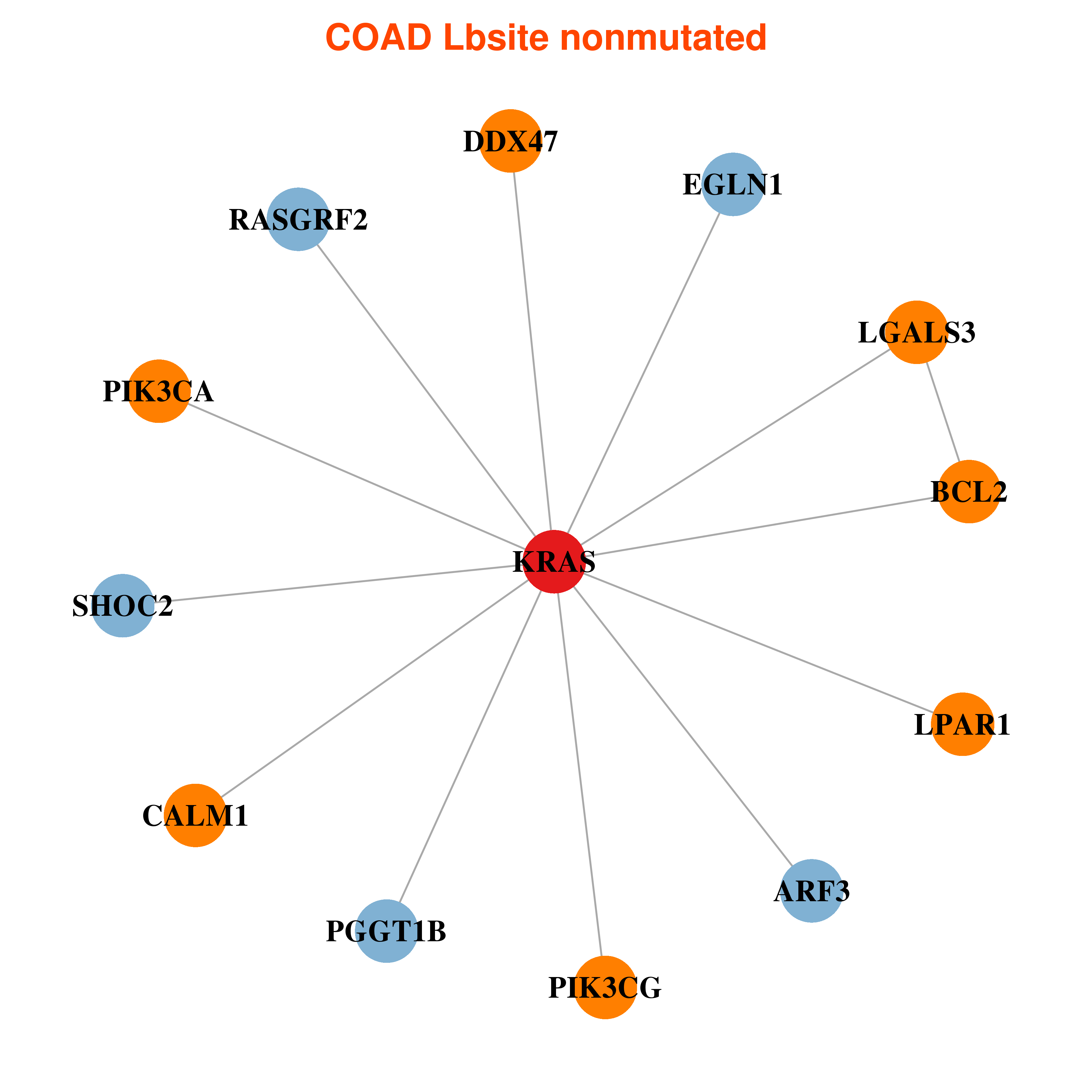
| KRAS_COAD_DE |
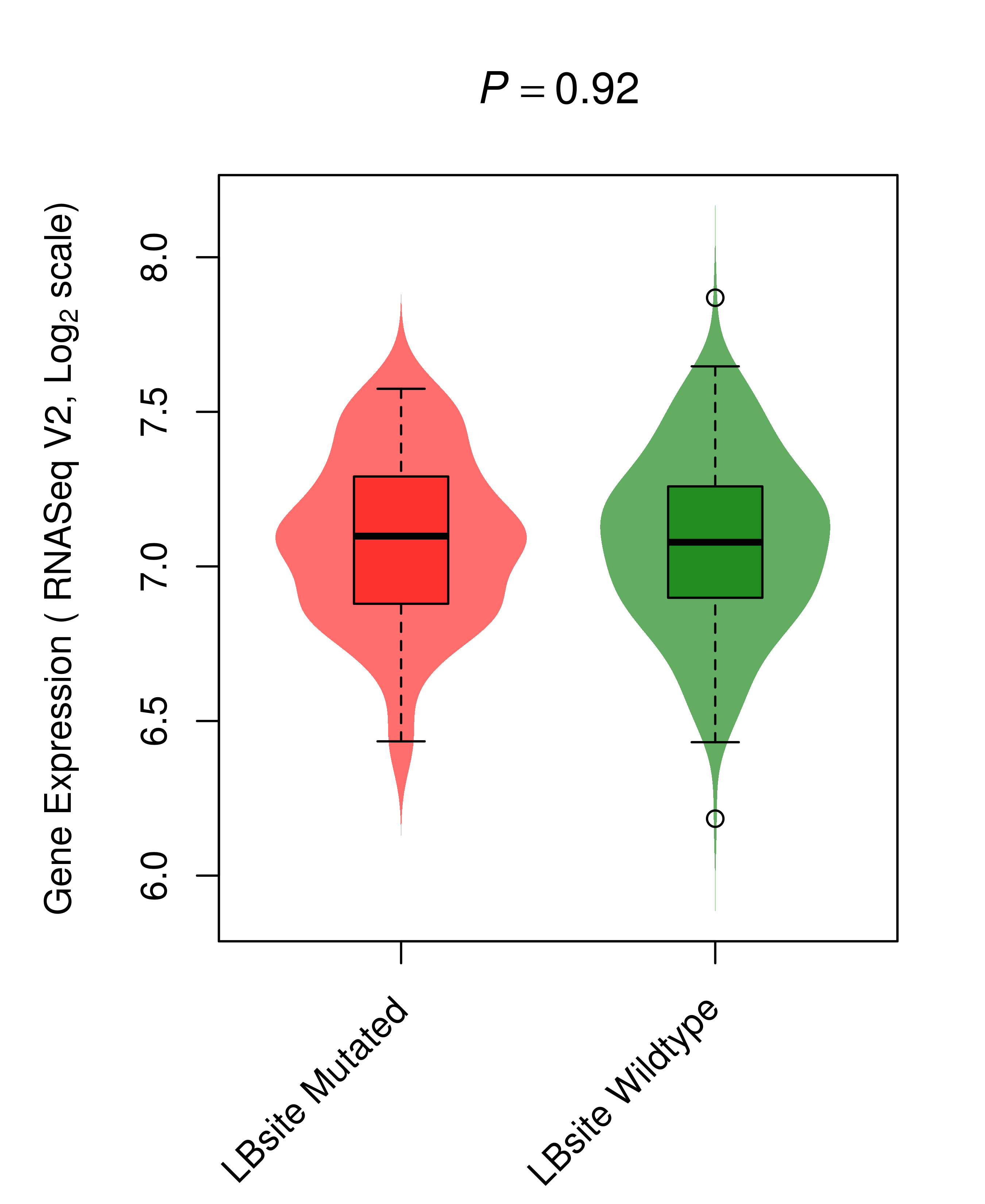 |
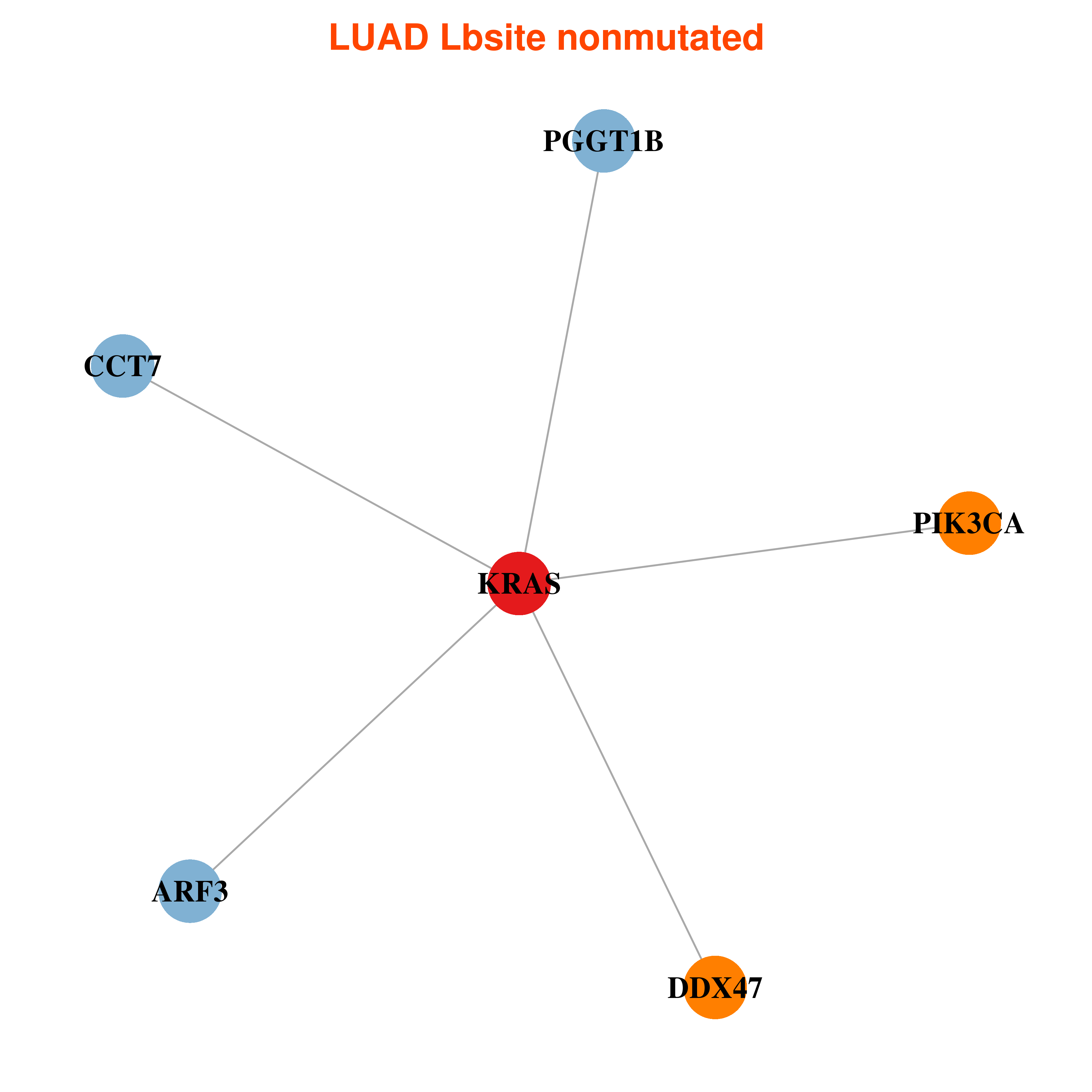
| KRAS_LUAD_DE |
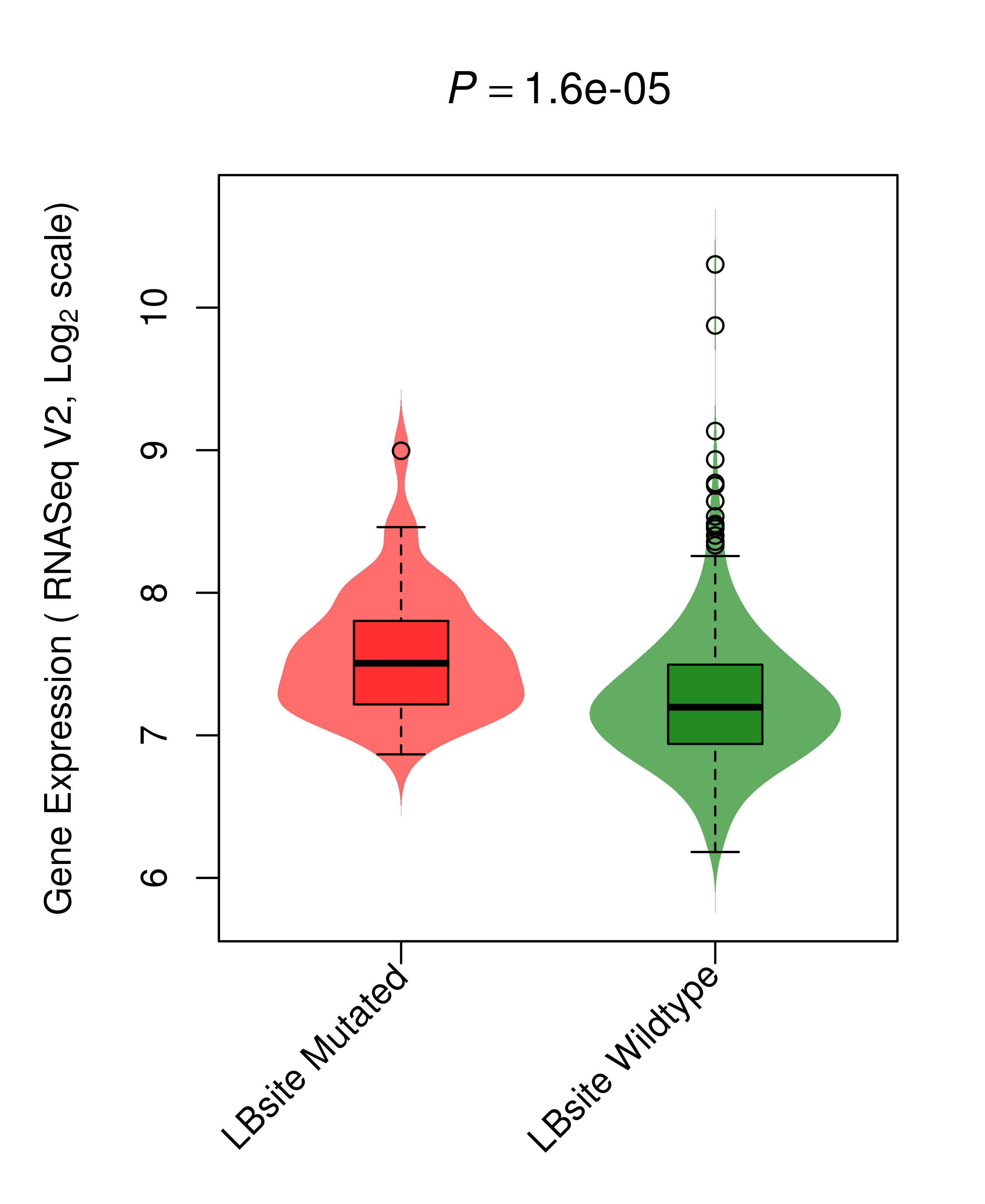 |
 Differential co-expressed gene network based on protein-protein interaction data (CePIN) Differential co-expressed gene network based on protein-protein interaction data (CePIN) |
| * In COAD | |
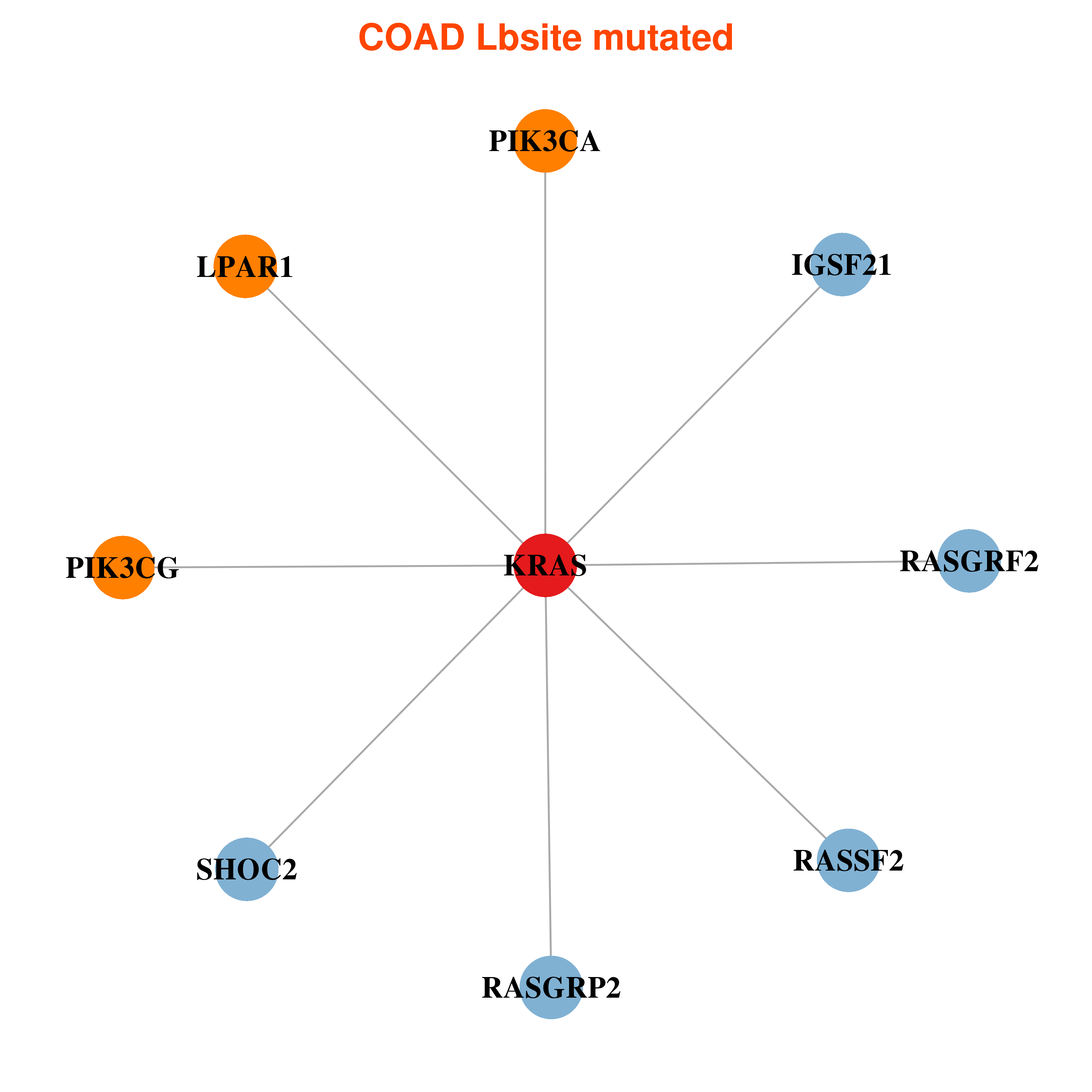 |
|
| * In LUAD | |
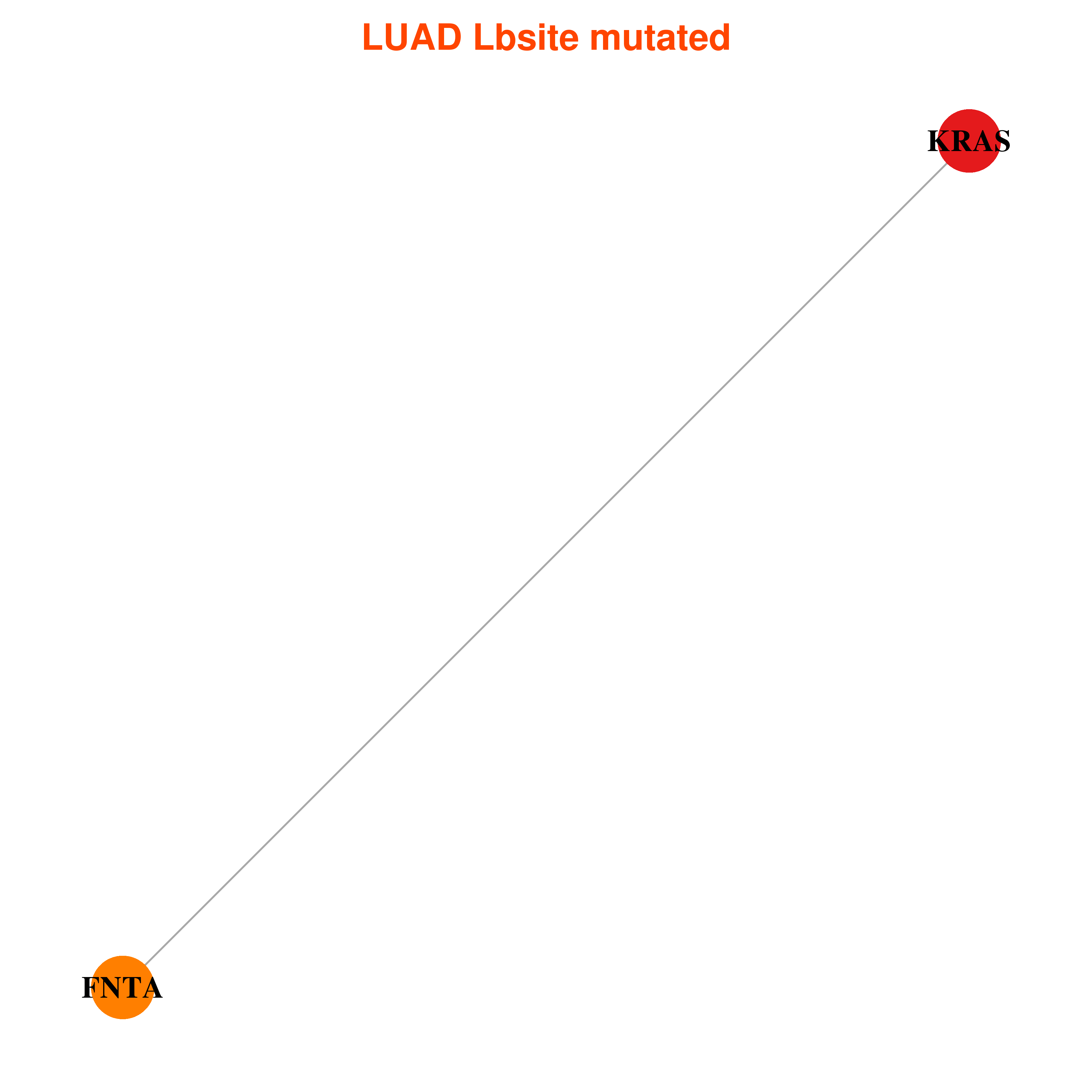 |
|
| Top |
| Top |
| Phenotype information for KRAS |
 Gene level disease information (DisGeNet) Gene level disease information (DisGeNet) |
| Disease ID | Disease name | # PubMed | Association type |
| umls:C0001418 | Adenocarcinoma | 228 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0007131 | Carcinoma, Non-Small-Cell Lung | 178 | Biomarker, GeneticVariation |
| umls:C0007097 | Carcinoma | 117 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0027627 | Neoplasm Metastasis | 100 | Biomarker, GeneticVariation |
| umls:C0024121 | Lung Neoplasms | 94 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0009404 | Colorectal Neoplasms | 92 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0152013 | Adenocarcinoma of lung | 73 | Biomarker, GeneticVariation |
| umls:C0001430 | Adenoma | 69 | Biomarker, GeneticVariation |
| umls:C0027651 | Neoplasms | 52 | Biomarker, GeneticVariation |
| umls:C0009375 | Colonic Neoplasms | 38 | Biomarker, GeneticVariation |
| umls:C0007137 | Carcinoma, Squamous Cell | 36 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0030297 | Pancreatic Neoplasms | 35 | Biomarker, GeneticVariation |
| umls:C0028326 | Noonan Syndrome | 32 | Biomarker, GeneticVariation |
| umls:C0919267 | Ovarian Neoplasms | 20 | Biomarker, GeneticVariation |
| umls:C0699791 | STOMACH CARCINOMA | 19 | Biomarker, GeneticVariation |
| umls:C0206698 | Cholangiocarcinoma | 18 | Biomarker, GeneticVariation |
| umls:C1275081 | Cardiofaciocutaneous syndrome | 15 | Biomarker |
| umls:C2239176 | Carcinoma, Hepatocellular | 12 | Biomarker |
| umls:C0040136 | Thyroid Neoplasms | 9 | Biomarker, GeneticVariation |
| umls:C0038356 | Stomach Neoplasms | 9 | Biomarker, GeneticVariation |
| umls:C0587248 | Costello Syndrome | 9 | Biomarker, GeneticVariation |
| umls:C0023467 | Leukemia, Myeloid, Acute | 8 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0014175 | Endometriosis | 7 | Biomarker, GeneticVariation |
| umls:C0023903 | Liver Neoplasms | 7 | Biomarker, GeneticVariation |
| umls:C0018923 | Hemangiosarcoma | 7 | Biomarker |
| umls:C1458155 | Breast Neoplasms | 4 | Biomarker |
| umls:C0005684 | Malignant neoplasm of bladder | 4 | Biomarker, GeneticVariation |
| umls:C0026640 | Mouth Neoplasms | 3 | Biomarker, GeneticVariation |
| umls:C0024299 | Lymphoma | 3 | Biomarker |
| umls:C0027659 | Neoplasms, Experimental | 3 | Biomarker |
| umls:C0005695 | Urinary Bladder Neoplasms | 2 | Biomarker, GeneticVariation |
| umls:C0004114 | Astrocytoma | 2 | Biomarker, GeneticVariation |
| umls:C2931822 | Nasopharyngeal carcinoma | 2 | Biomarker |
| umls:C0265318 | Nevus, Sebaceous of Jadassohn | 1 | Biomarker |
| umls:C0007528 | Cecal Neoplasms | 1 | Biomarker |
| umls:C0016978 | Gallbladder Neoplasms | 1 | Biomarker |
| umls:C0023897 | Liver Diseases, Parasitic | 1 | Biomarker |
| umls:C0023904 | Liver Neoplasms, Experimental | 1 | Biomarker |
| umls:C0030849 | Penile Neoplasms | 1 | Biomarker |
| umls:C0042138 | Uterine Neoplasms | 1 | Biomarker |
 Mutation level pathogenic information (ClinVar annotation) Mutation level pathogenic information (ClinVar annotation) |
| Allele ID | AA change | Clinical significance | Origin | Phenotype IDs |
| 27617 | G12C | Pathogenic;drug response | Somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0476089 OMIM:608089 SNOMED CT:254878006 MedGen:C0684249 OMIM:211980 SNOMED CT:187875007 |
| 27618 | G12R | Pathogenic;drug response | Somatic | MedGen:C0005684 OMIM:109800 SNOMED CT:399326009 MedGen:C0007131 SNOMED CT:254637007 MedGen:C0149782 |
| 27619 | G13D | Pathogenic;drug response | Somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0349639 OMIM:607785 Orphanet:ORPHA86834 MedGen:C0858252 MedGen:C2674723 OMIM:614470 Orphanet:ORPHA268114 |
| 27621 | G12D | Pathogenic;drug response | Somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0038356 OMIM:613659 SNOMED CT:126824007 MedGen:C0235974 OMIM:260350 Orphanet:ORPHA1333 SNOMED CT:372142002 MedGen:C0265318 OMIM:163200 Orphanet:ORPHA2612 SNOMED CT:239112008 MedGen:C0334082 OMIM:162900 Orphanet: |
| 27622 | G12V | Pathogenic;drug response | Germline;somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0235974 OMIM:260350 Orphanet:ORPHA1333 SNOMED CT:372142002 MedGen:C0349639 OMIM:607785 Orphanet:ORPHA86834 MedGen:C0853032 MedGen:CN166718 |
| 27623 | G12S | Pathogenic;drug response | Somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0038356 OMIM:613659 SNOMED CT:126824007 MedGen:C0349639 OMIM:607785 Orphanet:ORPHA86834 MedGen:C0919267 OMIM:167000 SNOMED CT:123843001 |
| 48924 | P34L | Pathogenic | Germline | GeneReviews:NBK1124 MedGen:C1860991 OMIM:609942 Orphanet:ORPHA648 MedGen:CN166718 MedGen:CN221809 |
| 54282 | Q61R | Pathogenic | Germline | GeneReviews:NBK1124 MedGen:C1860991 OMIM:609942 Orphanet:ORPHA648 |
| 54283 | Q61L | Likely pathogenic;Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0238462 SNOMED CT:255032005 |
| 54284 | Q61H | Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| 54289 | G12A | Pathogenic;drug response | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| 54290 | G13C | Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C2674723 OMIM:614470 Orphanet:ORPHA268114 |
| 54291 | G13V | Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| 175715 | Q61K | Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| 175716 | I36M | Likely pathogenic | Germline | GeneReviews:NBK1124 MedGen:C1860991 OMIM:609942 Orphanet:ORPHA648 |
| 175854 | Q61H | Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| 194404 | A146T | Uncertain significance | Germline | MedGen:CN221809 |
| Top |
| Pharmacological information for KRAS |
 Gene expression profile of anticancer drug treated cell-lines (CCLE) Gene expression profile of anticancer drug treated cell-lines (CCLE)Heatmap showing the correlation between gene expression and drug response across all the cell-lines. We chose the top 20 among 138 drugs.We used Pearson's correlation coefficient. |
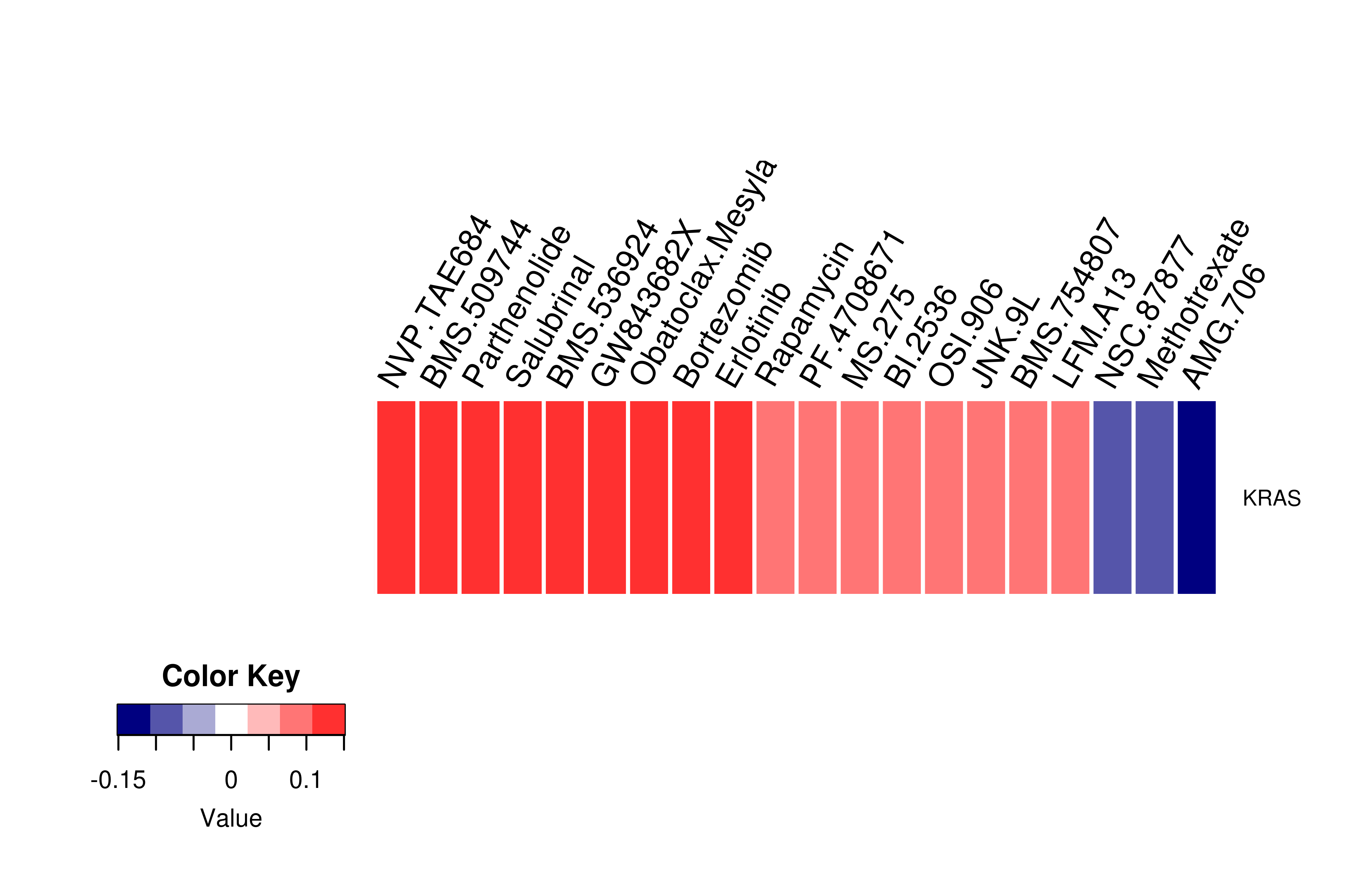 |
 Gene-centered drug-gene interaction network Gene-centered drug-gene interaction network |
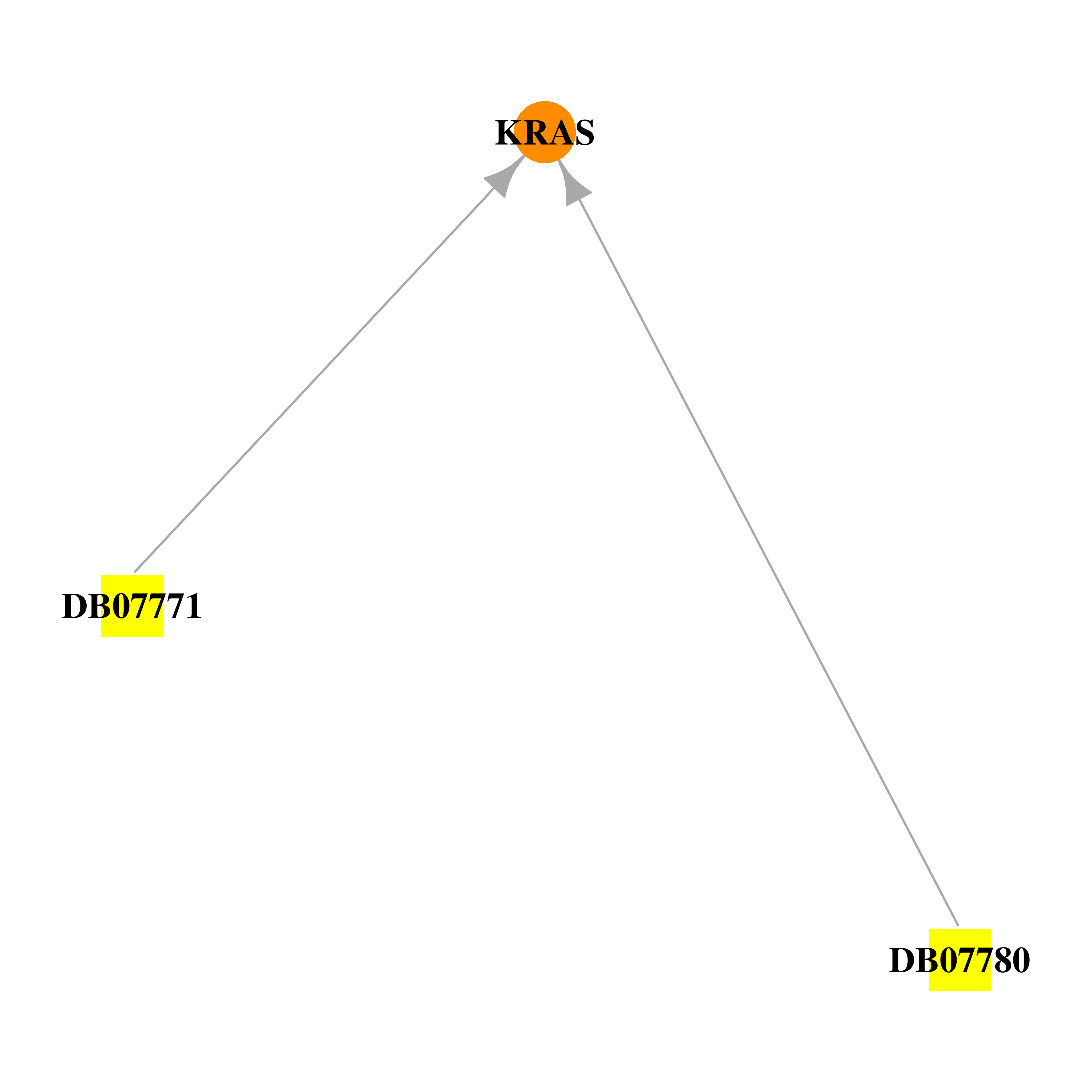 |
 Drug information targeting mutLBSgene (Approved drugs only) Drug information targeting mutLBSgene (Approved drugs only) |
| Drug status | DrugBank ID | Name | Type | Drug structure |
| Experimental | DB07771 | [(3,7,11-TRIMETHYL-DODECA-2,6,10-TRIENYLOXYCARBAMOYL)-METHYL]-PHOSPHONIC ACID | Small molecule | 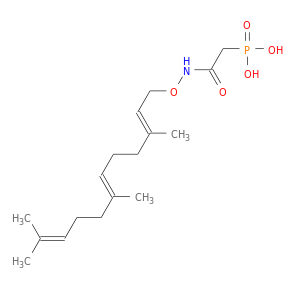 |
| Experimental | DB07780 | FARNESYL DIPHOSPHATE | Small molecule | 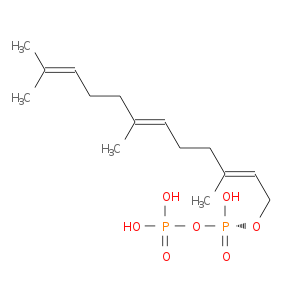 |
 Gene-centered ligand-gene interaction network Gene-centered ligand-gene interaction network |
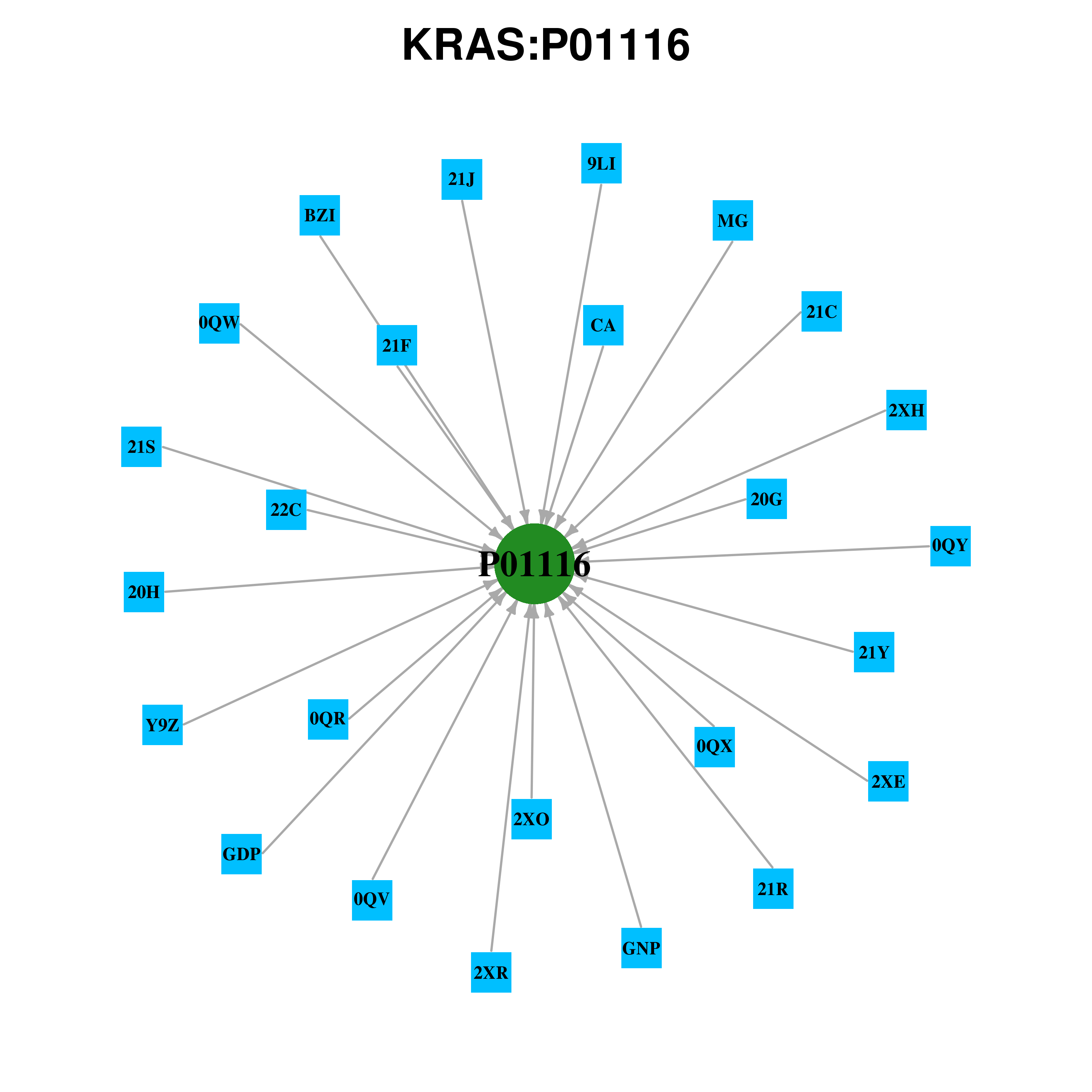 |
 Ligands binding to mutated ligand binding site of KRAS go to BioLip Ligands binding to mutated ligand binding site of KRAS go to BioLip |
| Ligand ID | Ligand short name | Ligand long name | PDB ID | PDB name | mutLBS | MG | MAGNESIUM(2+) | 4dsn | A | A59 | MG | MAGNESIUM(2+) | 4dst | A | A59 | 21F | N-{1-[N-(4-CHLORO-5-IODO-2-METHOXYPHENYL) GLYCYL]PIPERIDIN-4-YL}ETHANESULFONAMIDE | 4lyh | C | A59 R68 Y71 M72 | 21C | N-{1-[N-(4,5-DICHLORO-2-HYDROXYPHENYL)GLYCYL]PIPERIDIN- 4-YL}ETHANESULFONAMIDE | 4lyf | B | E62 R68 Y71 M72 | 21F | N-{1-[N-(4-CHLORO-5-IODO-2-METHOXYPHENYL) GLYCYL]PIPERIDIN-4-YL}ETHANESULFONAMIDE | 4lyj | A | E62 R68 Y71 M72 | 21J | N-(1-{[(5,7-DICHLORO-2,2-DIMETHYL-1,3-BENZODIOXOL-4- YL)OXY]ACETYL}PIPERIDIN-4-YL)ETHANESULFONAMIDE | 4m1o | B | E62 R68 Y71 M72 | 21R | N-{1-[N-(4,5-DICHLORO-2-ETHYLPHENYL)GLYCYL]PIPERIDIN-4- YL}ETHANESULFONAMIDE | 4m1w | B | E62 R68 Y71 M72 | GNP | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | 3gft | F | G12 G13 V14 S17 A18 F28 A59 K117 D119 A146 | GNP | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | 3gft | D | G12 G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4epw | A | G12 G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lpk | B | G12 G13 V14 S17 A18 F28 K117 D119 A146 | GNP | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | 3gft | C | G12 G13 V14 S17 A18 F28 T35 K117 D119 A146 | GNP | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | 3gft | A | G12 G13 V14 S17 A18 F28 Y32 P34 T35 K117 D119 A146 | GNP | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | 3gft | B | G12 G13 V14 S17 A18 F28 Y32 P34 T35 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lyf | C | G13 V14 S17 A18 F28 A59 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lyh | C | G13 V14 S17 A18 F28 A59 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1s | C | G13 V14 S17 A18 F28 A59 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1w | C | G13 V14 S17 A18 F28 A59 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1y | C | G13 V14 S17 A18 F28 A59 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m21 | C | G13 V14 S17 A18 F28 A59 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4dsu | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4epr | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4ept | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4epv | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4epx | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4epy | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4l8g | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lpk | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lrw | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4luc | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4luc | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lv6 | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lv6 | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lyf | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lyf | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lyh | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lyh | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1o | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1o | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1o | C | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1s | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1s | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1t | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1t | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1t | C | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1w | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1w | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1y | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m1y | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m21 | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m21 | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m22 | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m22 | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4m22 | C | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4ldj | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4obe | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4obe | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4pzy | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4pzy | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4pzz | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4q01 | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4q01 | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4q02 | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4q03 | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4ql3 | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4tq9 | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4tq9 | B | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4wa7 | A | G13 V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 5f2e | A | G13 V14 S17 A18 F28 K117 D119 A146 | Y9Z | 5'-O-[(S)-{[(S)-[2-(ACETYLAMINO)ETHOXY](HYDROXY) PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]GUANOSINE | 4nmm | A | G13 V14 S17 A18 F28 P34 A59 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lrw | B | G13 V14 S17 A18 F28 Y32 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4lyj | A | G13 V14 S17 A18 F28 Y32 K117 D119 A146 | GSP | GUANOSINE 5'-[GAMMA-THIO]TRIPHOSPHATE | 4dso | A | G13 V14 S17 A18 F28 Y32 P34 T35 K117 D119 A146 | GNP | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | 3gft | E | G13 V14 S17 A18 F28 Y32 T35 K117 D119 A146 | 9LI | 2-(4,6-DICHLORO-2-METHYL-1H-INDOL-3-YL)ETHANAMINE | 4dst | A | K5 V7 | BZI | BENZIMIDAZOLE | 4dsu | A | K5 V7 | 2XR | 2-CHLORO-1-(1H-INDOL-3-YL)ETHANONE | 4pzy | A | K5 V7 | 0QX | 2-(1H-INDOL-3-YLMETHYL)-1H-IMIDAZO[4,5-C]PYRIDINE | 4epv | A | K5 V7 E37 Y71 | 0QV | (4-HYDROXYPIPERIDIN-1-YL)(1H-INDOL-3-YL)METHANETHIONE | 4epw | A | K5 V7 E37 Y71 | 0QY | N-[2-(1H-INDOL-3-YLMETHYL)-1H-BENZIMIDAZOL-5-YL]-L- PROLINAMIDE | 4epy | A | K5 V7 E37 Y71 | 0QW | (2-HYDROXYPHENYL)(PYRROLIDIN-1-YL)METHANETHIONE | 4ept | A | K5 V7 Y71 | 2XR | 2-CHLORO-1-(1H-INDOL-3-YL)ETHANONE | 4pzy | B | K5 V7 Y71 | 2XO | 1H-BENZIMIDAZOL-2-YLMETHANETHIOL | 4pzz | A | K5 V7 Y71 | 2XH | NAPHTHALENE-1-THIOL | 4q01 | A | K5 V7 Y71 | 2XH | NAPHTHALENE-1-THIOL | 4q01 | B | K5 V7 Y71 | 2XG | 3,4-DIFLUOROBENZENETHIOL | 4q02 | A | K5 V7 Y71 | 5UT | 1-[3-[4-[2-[[4-CHLORANYL-5-(1-METHYLCYCLOPROPYL)-2- OXIDANYL-PHENYL]AMINO]ETHANOYL]PIPERAZIN-1- YL]AZETIDIN-1-YL]PROP-2-EN-1-ONE | 5f2e | A | P34 A59 E62 R68 M72 | 22C | 1-{4-[(2,4-DICHLOROPHENOXY)ACETYL]PIPERAZIN-1- YL}PROPAN-1-ONE | 4m22 | B | P34 A59 R68 Y71 M72 | 22C | 1-{4-[(2,4-DICHLOROPHENOXY)ACETYL]PIPERAZIN-1- YL}PROPAN-1-ONE | 4m22 | C | P34 A59 Y71 M72 | 21Y | 1-(4-{[(4,5-DICHLORO-2-METHOXYPHENYL) AMINO]ACETYL}PIPERAZIN-1-YL)PROPAN-1-ONE | 4m21 | B | R68 Y71 M72 | MG | MAGNESIUM(2+) | 3gft | D | S17 | MG | MAGNESIUM(2+) | 3gft | A | S17 T35 | MG | MAGNESIUM(2+) | 3gft | B | S17 T35 | MG | MAGNESIUM(2+) | 3gft | C | S17 T35 | MG | MAGNESIUM(2+) | 3gft | E | S17 T35 | MG | MAGNESIUM(2+) | 4dsn | A | S17 T35 | MG | MAGNESIUM(2+) | 4dso | A | S17 T35 | MG | MAGNESIUM(2+) | 4dst | A | S17 T35 | CA | CALCIUM(2+) | 4lpk | B | S17 Y32 | MG | MAGNESIUM(2+) | 4lyj | A | S17 Y32 | MG | MAGNESIUM(2+) | 4nmm | A | S17 Y32 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4tqa | A | V14 S17 A18 F28 K117 D119 A146 | GDP | GUANOSINE-5'-DIPHOSPHATE | 4tqa | B | V14 S17 A18 F28 K117 D119 A146 | 21R | N-{1-[N-(4,5-DICHLORO-2-ETHYLPHENYL)GLYCYL]PIPERIDIN-4- YL}ETHANESULFONAMIDE | 4m1w | C | V7 A59 Q61 Y71 M72 | 0QR | N-(6-AMINOPYRIDIN-2-YL)-4-FLUOROBENZENESULFONAMIDE | 4epx | A | V7 E37 E62 Q70 Y71 | 21F | N-{1-[N-(4-CHLORO-5-IODO-2-METHOXYPHENYL) GLYCYL]PIPERIDIN-4-YL}ETHANESULFONAMIDE | 4lyh | B | V7 E62 R68 Y71 M72 | 21M | N-{1-[(2,4-DICHLOROPHENOXY)ACETYL]PIPERIDIN-4- YL}ETHANESULFONAMIDE | 4m1t | B | V7 E62 R68 Y71 M72 | 21S | N-{1-[N-(5,7-DICHLORO-2,1,3-BENZOTHIADIAZOL-4-YL) GLYCYL]PIPERIDIN-4-YL}ETHANESULFONAMIDE | 4m1y | B | V7 E62 R68 Y71 M72 | 20G | N-{1-[(2,4-DICHLOROPHENOXY)ACETYL]PIPERIDIN-4-YL}-4- SULFANYLBUTANAMIDE | 4luc | A | V7 G13 P34 Q61 E62 R68 Y71 M72 | 20H | 1-[(2,4-DICHLOROPHENOXY)ACETYL]-N-(2-SULFANYLETHYL) PIPERIDINE-4-CARBOXAMIDE | 4lv6 | A | V7 P34 A59 Q61 E62 R68 Y71 M72 | 20H | 1-[(2,4-DICHLOROPHENOXY)ACETYL]-N-(2-SULFANYLETHYL) PIPERIDINE-4-CARBOXAMIDE | 4lv6 | B | V7 P34 A59 Q61 E62 R68 Y71 M72 | 20G | N-{1-[(2,4-DICHLOROPHENOXY)ACETYL]PIPERIDIN-4-YL}-4- SULFANYLBUTANAMIDE | 4luc | B | V7 Q61 E62 R68 Y71 M72 | 21K | N-{1-[N-(2,4-DICHLOROPHENYL)GLYCYL]PIPERIDIN-4- YL}ETHANESULFONAMIDE | 4m1s | B | V7 Q61 R68 Y71 M72 | 2XE | 4-BROMOBENZENETHIOL | 4q03 | A | V7 Y71 |
| Top |
| Conservation information for LBS of KRAS |
 Multiple alignments for P01116 in multiple species Multiple alignments for P01116 in multiple species |
| LBS | AA sequence | # species | Species | A146 | FIETSAKTRQR | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | A146 | YIETSAKTRMG | 1 | Drosophila melanogaster | A146 | NVDTSAKTRMG | 1 | Caenorhabditis elegans | A18 | GVGKSALTIQL | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | A59 | DILDTAGQEEY | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | D119 | VGNKCDLPSRT | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | D119 | VGNKCDLASWN | 1 | Drosophila melanogaster | D119 | VGNKCDLSSRS | 1 | Caenorhabditis elegans | D30 | QNHFVDEYDPT | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | D30 | QNHFVEEYDPT | 1 | Caenorhabditis elegans | D38 | DPTIEDSYRKQ | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | D54 | ETCLLDILDTA | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | E31 | NHFVDEYDPTI | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | E31 | NHFVEEYDPTI | 1 | Caenorhabditis elegans | E37 | YDPTIEDSYRK | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | E62 | DTAGQEEYSAM | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | F28 | LIQNHFVDEYD | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | F28 | LIQNHFVEEYD | 1 | Caenorhabditis elegans | G12 | VVVGAGGVGKS | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | G12 | VVVGDGGVGKS | 1 | Caenorhabditis elegans | G13 | VVGAGGVGKSA | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | G13 | VVGDGGVGKSA | 1 | Caenorhabditis elegans | G15 | GAGGVGKSALT | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | G15 | GDGGVGKSALT | 1 | Caenorhabditis elegans | G60 | ILDTAGQEEYS | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | G75 | QYMRTGEGFLC | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | G75 | QYMRTGEGFLL | 2 | Drosophila melanogaster, Caenorhabditis elegans | I100 | HYREQIKRVKD | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | I100 | TYREQIKRVKD | 1 | Drosophila melanogaster | I100 | NYREQIRRVKD | 1 | Caenorhabditis elegans | I55 | TCLLDILDTAG | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | K117 | VLVGNKCDLPS | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | K117 | VLVGNKCDLAS | 1 | Drosophila melanogaster | K117 | VLVGNKCDLSS | 1 | Caenorhabditis elegans | K16 | AGGVGKSALTI | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | K16 | DGGVGKSALTI | 1 | Caenorhabditis elegans | K5 | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | L120 | GNKCDLPSRTV | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | L120 | GNKCDLASWNV | 1 | Drosophila melanogaster | L120 | GNKCDLSSRSV | 1 | Caenorhabditis elegans | L56 | CLLDILDTAGQ | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | L6 | MTEYKLVVVGA | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | L6 | MTEYKLVVVGD | 1 | Caenorhabditis elegans | M67 | EEYSAMRDQYM | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | M72 | MRDQYMRTGEG | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | N116 | MVLVGNKCDLP | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | N116 | MVLVGNKCDLA | 1 | Drosophila melanogaster | N116 | MVLVGNKCDLS | 1 | Caenorhabditis elegans | P34 | VDEYDPTIEDS | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | P34 | VEEYDPTIEDS | 1 | Caenorhabditis elegans | Q61 | LDTAGQEEYSA | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | Q70 | SAMRDQYMRTG | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | Q99 | HHYREQIKRVK | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | Q99 | GTYREQIKRVK | 1 | Drosophila melanogaster | Q99 | ANYREQIRRVK | 1 | Caenorhabditis elegans | R68 | EYSAMRDQYMR | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | R73 | RDQYMRTGEGF | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | S145 | PFIETSAKTRQ | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | S145 | PYIETSAKTRM | 1 | Drosophila melanogaster | S145 | PNVDTSAKTRM | 1 | Caenorhabditis elegans | S17 | GGVGKSALTIQ | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | S39 | PTIEDSYRKQV | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | T35 | DEYDPTIEDSY | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | T35 | EEYDPTIEDSY | 1 | Caenorhabditis elegans | T58 | LDILDTAGQEE | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | T74 | DQYMRTGEGFL | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | V14 | VGAGGVGKSAL | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | V14 | VGDGGVGKSAL | 1 | Caenorhabditis elegans | V29 | IQNHFVDEYDP | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | V29 | IQNHFVEEYDP | 1 | Caenorhabditis elegans | V7 | TEYKLVVVGAG | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | V7 | TEYKLVVVGDG | 1 | Caenorhabditis elegans | Y32 | HFVDEYDPTIE | 4 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster | Y32 | HFVEEYDPTIE | 1 | Caenorhabditis elegans | Y40 | TIEDSYRKQVV | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | Y71 | AMRDQYMRTGE | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans | Y96 | EDIHHYREQIK | 3 | Homo sapiens, Mus musculus, Rattus norvegicus | Y96 | EDIGTYREQIK | 1 | Drosophila melanogaster | Y96 | ENVANYREQIR | 1 | Caenorhabditis elegans |
 |
Copyright © 2016-Present - The University of Texas Health Science Center at Houston |