|
mutLBSgeneDB |
| |
| |
| |
| |
| |
| |
|
| Gene summary for BRAF |
 Gene summary Gene summary |
| Basic gene Info. | Gene symbol | BRAF |
| Gene name | B-Raf proto-oncogene, serine/threonine kinase | |
| Synonyms | B-RAF1|BRAF1|NS7|RAFB1 | |
| Cytomap | UCSC genome browser: 7q34 | |
| Type of gene | protein-coding | |
| RefGenes | NM_004333.4, | |
| Description | 94 kDa B-raf proteinB-Raf proto-oncogene serine/threonine-protein kinase (p94)murine sarcoma viral (v-raf) oncogene homolog B1proto-oncogene B-Rafserine/threonine-protein kinase B-rafv-raf murine sarcoma viral oncogene homolog Bv-raf murine sarcoma | |
| Modification date | 20141222 | |
| dbXrefs | MIM : 164757 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000157764 | ||
| HPRD : 01264 | ||
| Vega : OTTHUMG00000157457 | ||
| Protein | UniProt: P15056 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_BRAF | |
| BioGPS: 673 | ||
| Pathway | NCI Pathway Interaction Database: BRAF | |
| KEGG: BRAF | ||
| REACTOME: BRAF | ||
| Pathway Commons: BRAF | ||
| Context | iHOP: BRAF | |
| ligand binding site mutation search in PubMed: BRAF | ||
| UCL Cancer Institute: BRAF | ||
| Assigned class in mutLBSgeneDB | A: This gene has a literature evidence and it belongs to targetable_mutLBSgenes. | |
| References showing study about ligand binding site mutation for BRAF. | 1. "Kondo T, Nakazawa T, Murata S, Kurebayashi J, Ezzat S, Asa SL, Katoh R. Enhanced B-Raf protein expression is independent of V600E mutant status in thyroid carcinomas. Hum Pathol. 2007 Dec;38(12):1810-8. Epub 2007 Aug 21. PubMed PMID: 17714762. " 17714762 2. "Angell TE, Lechner MG, Jang JK, Correa AJ, LoPresti JS, Epstein AL. BRAF V600E in papillary thyroid carcinoma is associated with increased programmed death ligand 1 expression and suppressive immune cell infiltration. Thyroid. 2014 Sep;24(9):1385-93. doi: 10.1089/thy.2014.0134. Epub 2014 Jul 15. PubMed PMID: 24955518; PubMed Central PMCID: PMC4148060." 24955518 3. "Tang HC, Chen YC. Insight into molecular dynamics simulation of BRAF(V600E) and potent novel inhibitors for malignant melanoma. Int J Nanomedicine. 2015 Apr 23;10:3131-46. doi: 10.2147/IJN.S80150. eCollection 2015. PubMed PMID: 25960652; PubMed Central PMCID: PMC4412490. " 25960652 | |
 Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez |
| GO ID | GO Term | PubMed ID | GO:0006468 | protein phosphorylation | 17563371 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 19667065 | GO:0043066 | negative regulation of apoptotic process | 19667065 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 22065586 | GO:0071277 | cellular response to calcium ion | 18567582 |
| Top |
| Ligand binding site mutations for BRAF |
 Lollipop-style diagram of mutations at LBS in amino-acid sequence. Lollipop-style diagram of mutations at LBS in amino-acid sequence. We represented ligand binding site mutations only. (You can see big image via clicking.) |
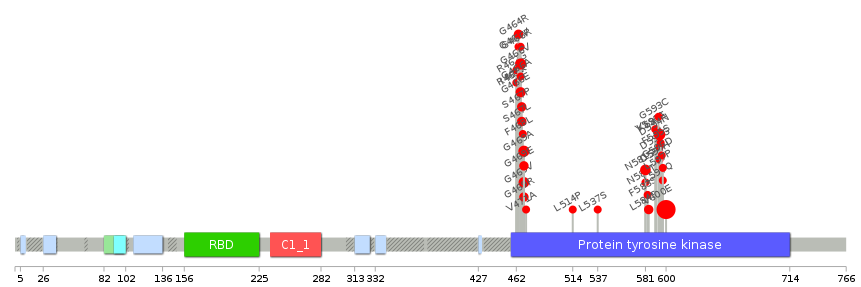 |
 Cancer type specific mutLBS sorted by frequency Cancer type specific mutLBS sorted by frequency |
| LBS | AAchange of nsSNV | Cancer type | # samples | A598 | V600E | THCA | 182 | A598 | V600E | SKCM | 85 | A598 | V600E | COAD | 20 | A598 | V600E | GBM | 5 | F468 | G469V | LUAD | 4 | D594 | D594N | COAD | 3 | G466 | G466V | LUAD | 3 | G466 | G466E | SKCM | 3 | G464 | G464R | COAD | 2 | F468,G466 | S467P | COAD | 2 | F468 | G469A | COAD | 2 | F583 | L584P | COAD | 2 | N581 | N581S | LUAD | 2 | A598 | V600E | LUAD | 2 | F468 | G469A | LUAD | 2 | F468,G466 | S467L | SKCM | 2 | Y538,S536 | L537S | BRCA | 1 | G593 | G593C | COAD | 1 | D594 | D594G | COAD | 1 | F583 | F583S | COAD | 1 | I463 | R462G | COAD | 1 | L597 | L597P | COAD | 1 | I592 | K591E | COAD | 1 | I592 | V590A | COAD | 1 | V471 | V471A | COAD | 1 | I463 | I463V | COAD | 1 | G466 | G466R | COAD | 1 | G466 | G466V | COAD | 1 | F468 | F468L | COAD | 1 | F468 | G469E | COAD | 1 | F595 | F595S | COAD | 1 | G596 | G596D | GBM | 1 | D594 | D594N | LUAD | 1 | L514 | L514P | LUAD | 1 | D594 | D594H | LUAD | 1 | G466 | G466A | LUAD | 1 | F468 | G469R | LUSC | 1 | G466 | G466V | LUSC | 1 | G464 | G464V | LUSC | 1 | N581 | N581S | OV | 1 | N581 | N581T | SKCM | 1 | F468 | G469A | SKCM | 1 | D594 | D594N | SKCM | 1 | L597 | L597Q | SKCM | 1 | F468 | G469R | SKCM | 1 | N581 | N581S | SKCM | 1 | F468 | G469E | SKCM | 1 | D594 | D594N | STAD | 1 | I463 | R462K | UCEC | 1 |
| cf) Cancer type abbreviation. BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, COAD: Colon adenocarcinoma, GBM: Glioblastoma multiforme, LGG: Brain lower grade glioma, HNSC: Head and neck squamous cell carcinoma, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute myeloid leukemia, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PRAD: Prostate adenocarcinoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, THCA: Thyroid carcinoma, UCEC: Uterine corpus endometrial carcinoma. |
 Clinical information for BRAF from My Cancer Genome. Clinical information for BRAF from My Cancer Genome. |
| The V600E mutation results in an amino acid substitution at position 600 in BRAF, from a valine (V) to a glutamic acid (E). This mutation occurs within the activation segment of the kinase domain (Figure 1). Approximately 80–90% of V600 BRAF mutations are V600E (COSMIC; Lovly et al. 2012; Rubinstein et al. 2010). Mutations at V600 result in increased kinase activity and are transforming in vitro. BRAF mutations are usually found in tumors with no driver mutations in NRAS, KIT, and other genes.BRAF V600E mutations are associated with increased sensitivity to BRAF inhibitors (Chapman et al. 2011; Falchook et al. 2012a; Flaherty et al. 2010; Flaherty et al. 2012a; Hauschild et al. 2012; Sosman et al. 2012).Patients whose tumors harbored V600E and V600K mutations showed better responses to the MEK inhibitor, trametinib, than to chemotherapy (dacarbazine or paclitaxel; Flaherty et al. 2012b); patients with V600E or V600K-mutated tumors also showed better responses to trametinib than patients with BRAF wild type tumors (Falchook et al. 2012b).The FDA has approved use of the combination therapy dabrafenib and trametinib for melanoma patients with V600E or V600K mutations based on an interim analysis of a phase III trial showing improved response rates and response duration compared to dabrafenib alone (FDA 2014; Long et al. 2014). Improved progression-free survival and response rates compared to dabrafenib monotherapy were observed in an earlier phase I/II trial, as well (Flaherty et al. 2012a).Patients with V600-mutated tumors and no prior BRAF inhibitors treated with combination vemurafenib and cobimetinib experienced improved response rate and progression-free survival compared to patients with V600-mutated tumors and recent progression on vemurafenib (Ribas et al. 2014). This study included patients with V600E and V600K mutations (Ribas et al. 2014).Lovly, C., W. Pao, J. Sosman. 2015. BRAF c.1799T>A (V600E) Mutation in Melanoma. My Cancer Genome https://www.mycancergenome.org/content/disease/melanoma/braf/54/ (Updated June 16). |
| Top |
| Protein structure related information for BRAF |
 Protein structure of wild type (WT) and mutant type (MT) of BRAF Protein structure of wild type (WT) and mutant type (MT) of BRAF |
| Wild type BRAF |
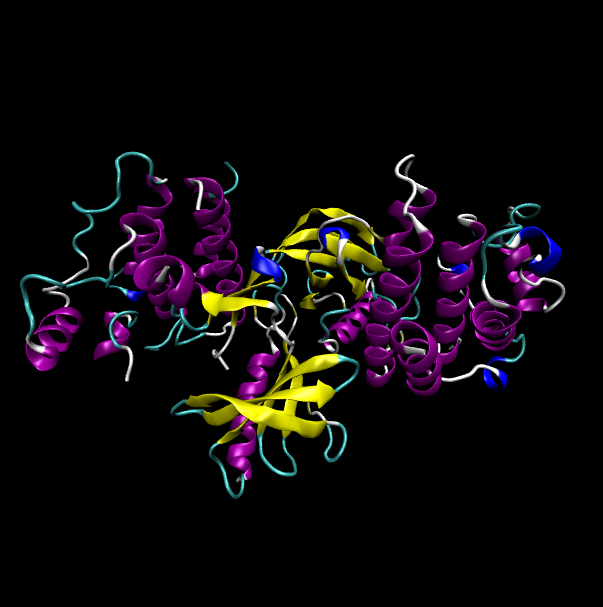 |
| Mutant type BRAF |
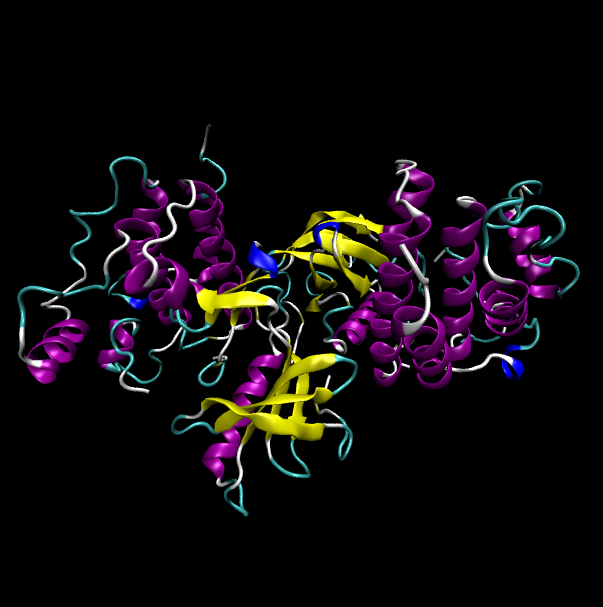 |
 Free energy of binding of drugs to wild type and mutant tpye of BRAF Free energy of binding of drugs to wild type and mutant tpye of BRAF |
| Gene symbol | Drug name | Free energy of binding (kcal/mol) of wild type | Free energy of binding (kcal/mol) of mutant type | BRAF | Dabrafenib | -10.1 | -9 | BRAF | Regorafenib | -9.7 | -9.4 | BRAF | Vemurafenib | -9.7 | -11.8 |
 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Mupro score denotes assessment of the effect of mutations on thermodynamic stability. (ΔΔE<0: mutation decreases stability, ΔΔE>0: mutation increases stability) |
 : nsSNV at non-LBS : nsSNV at non-LBS : nsSNV at LBS : nsSNV at LBS |
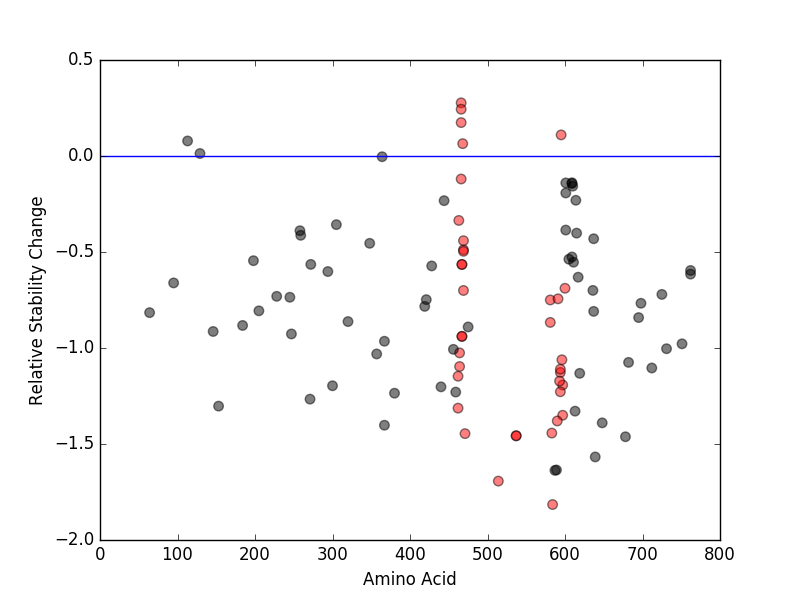 |
 nsSNVs sorted by the relative stability change of protein structure by each mutation nsSNVs sorted by the relative stability change of protein structure by each mutation Blue: mutations of positive stability change. and red : the most recurrent mutation for this gene. |
| LBS | AAchange of nsSNV | Relative stability change | G466 | G466V | 0.27685229 | G466 | G466E | 0.24351413 | G466 | G466R | 0.17410586 | F595 | F595S | 0.10990751 | F468 | F468L | 0.064337218 | F583 | L584P | -1.8152841 | L514 | L514P | -1.693253 | S536 | L537S | -1.4571753 | Y538 | L537S | -1.4571753 | V471 | V471A | -1.4459912 | F583 | F583S | -1.4431249 | I592 | V590A | -1.3800203 | L597 | L597Q | -1.3503094 | I463 | R462G | -1.313254 | D594 | D594G | -1.2279984 | L597 | L597P | -1.1926655 | G593 | G593C | -1.1724717 | I463 | R462K | -1.1470357 | D594 | D594N | -1.1272623 | D594 | D594H | -1.1104892 | G464 | G464R | -1.0964551 | G596 | G596D | -1.061375 | G464 | G464V | -1.0253723 | G466 | S467P | -0.93947437 | F468 | S467P | -0.93947437 | N581 | N581S | -0.86678303 | N581 | N581T | -0.74975157 | I592 | K591E | -0.7439051 | F468 | G469A | -0.70038964 | A598 | V600E | -0.68886259 | G466 | S467L | -0.56490356 | F468 | S467L | -0.56490356 | F468 | G469R | -0.49648081 | F468 | G469V | -0.48817585 | F468 | G469E | -0.44113949 | I463 | I463V | -0.33562603 | G466 | G466A | -0.11982765 |
| (MuPro1.1: Jianlin Cheng et al., Prediction of Protein Stability Changes for Single-Site Mutations Using Support Vector Machines, PROTEINS: Structure, Function, and Bioinformatics. 2006, 62:1125-1132) |
 Structure image for BRAF from PDB Structure image for BRAF from PDB |
| PDB ID | PDB title | PDB structure | 3OG7 | B-Raf Kinase V600E oncogeic mutant in complex with PLX4032 | 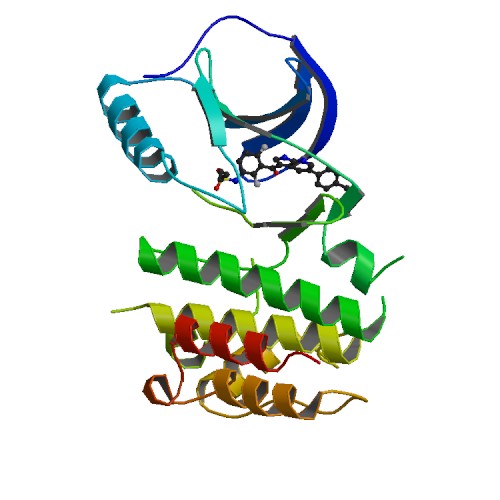 |
| Top |
| Differential gene expression and gene-gene network for BRAF |
 Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types |
| BRAF_COAD_DE |
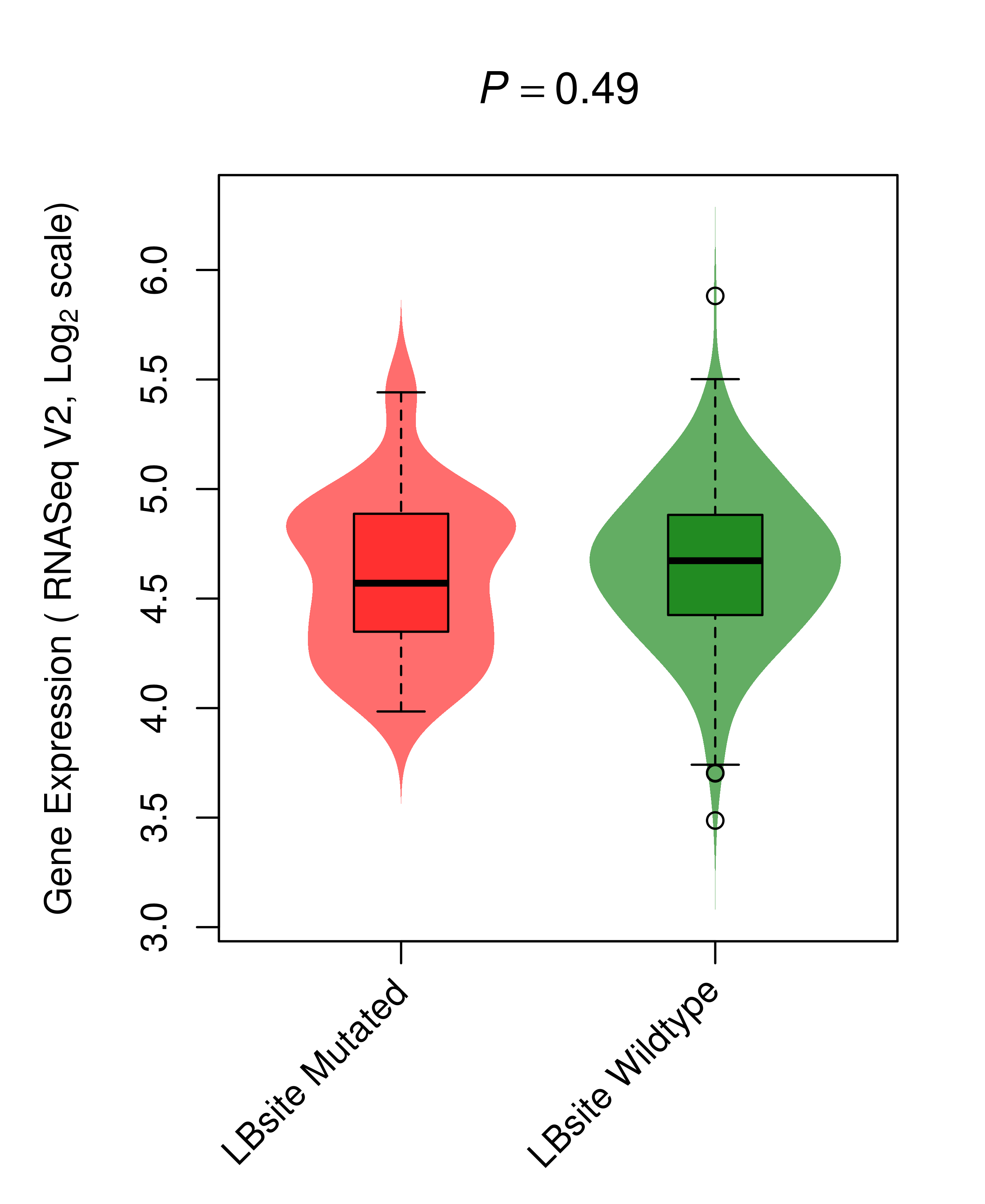 |
| BRAF_LUAD_DE |
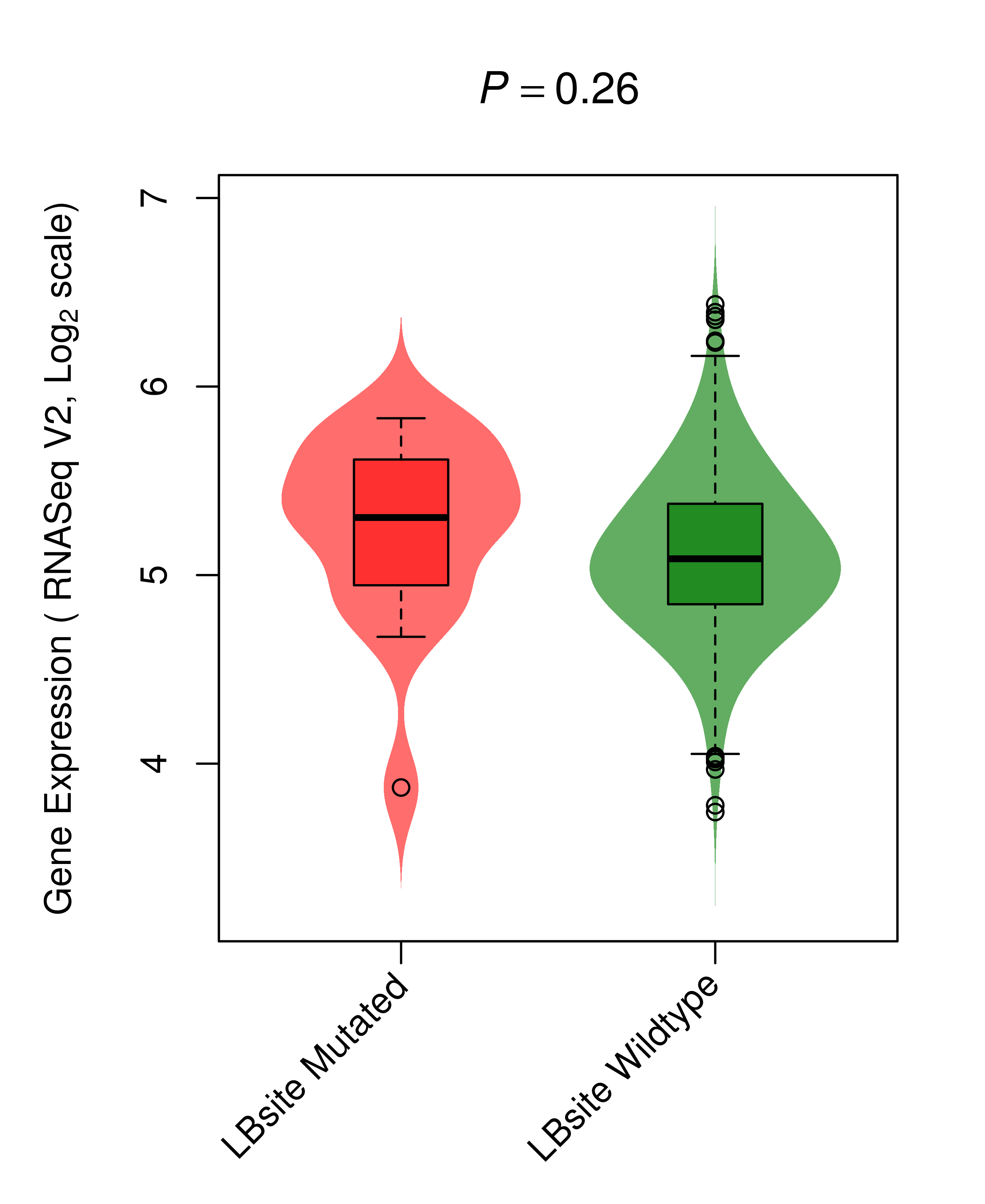 |
| BRAF_THCA_DE |
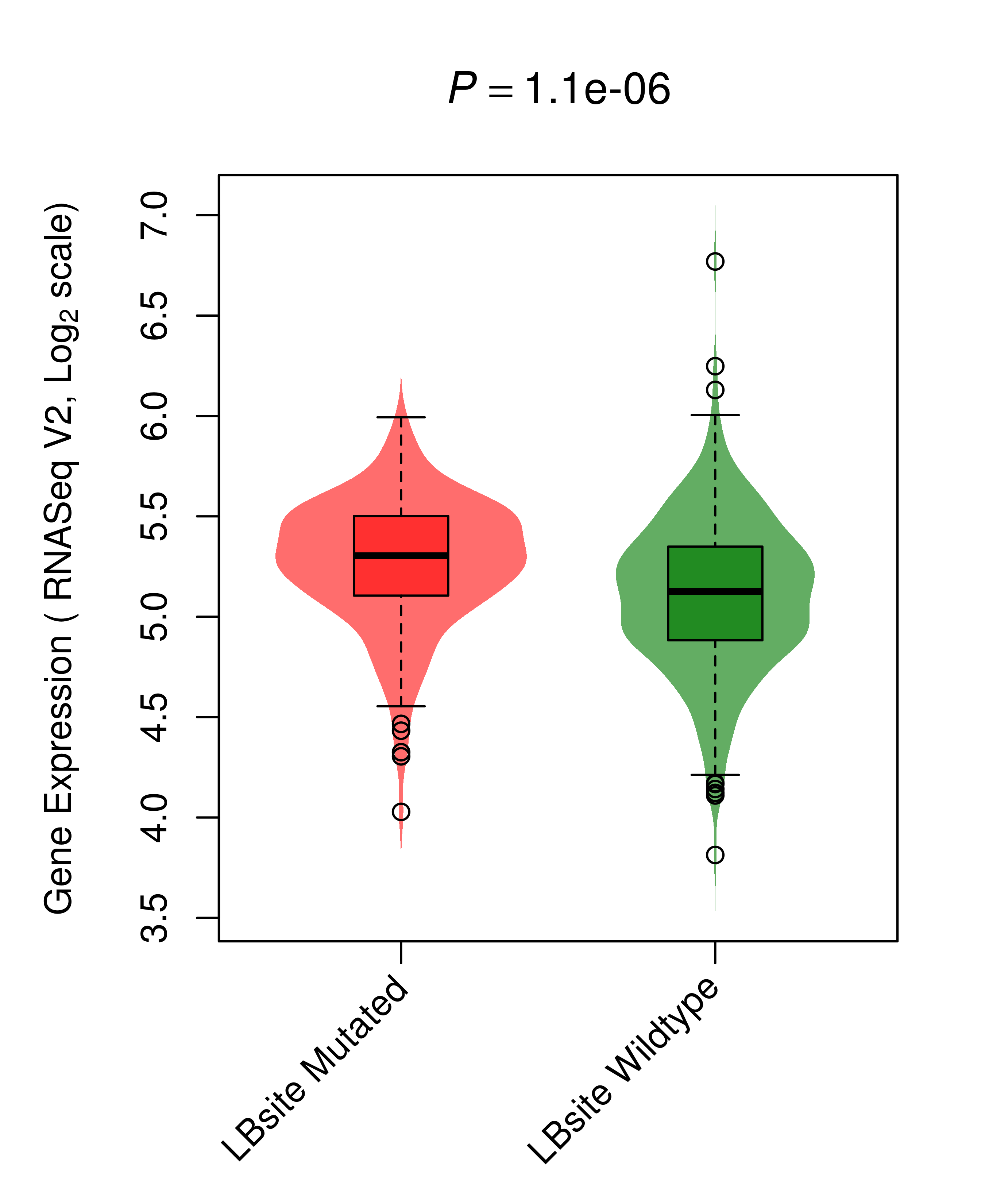 |
 Differential co-expressed gene network based on protein-protein interaction data (CePIN) Differential co-expressed gene network based on protein-protein interaction data (CePIN) |
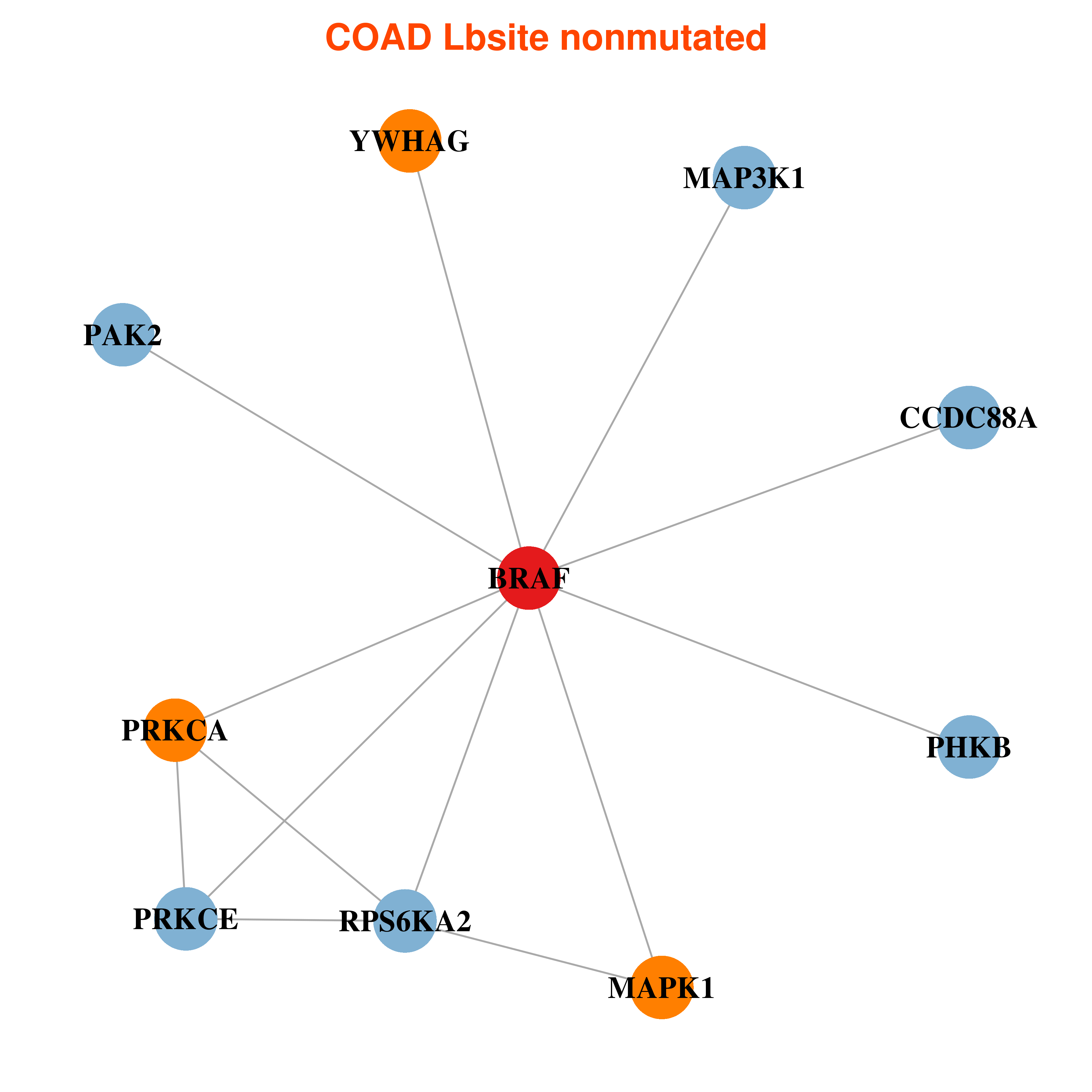
| * In COAD | |
 |
|
| * In LUAD | |
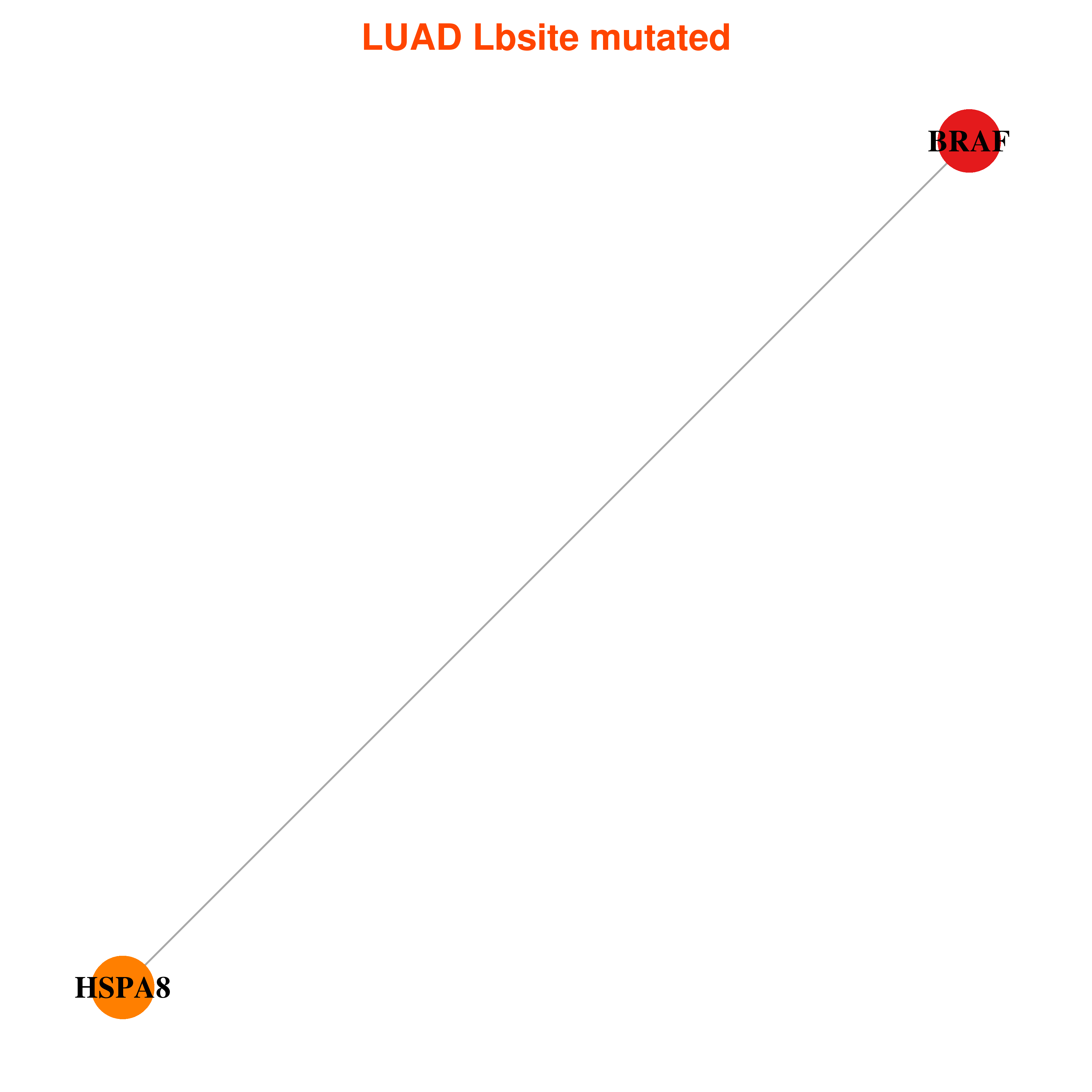 |
|
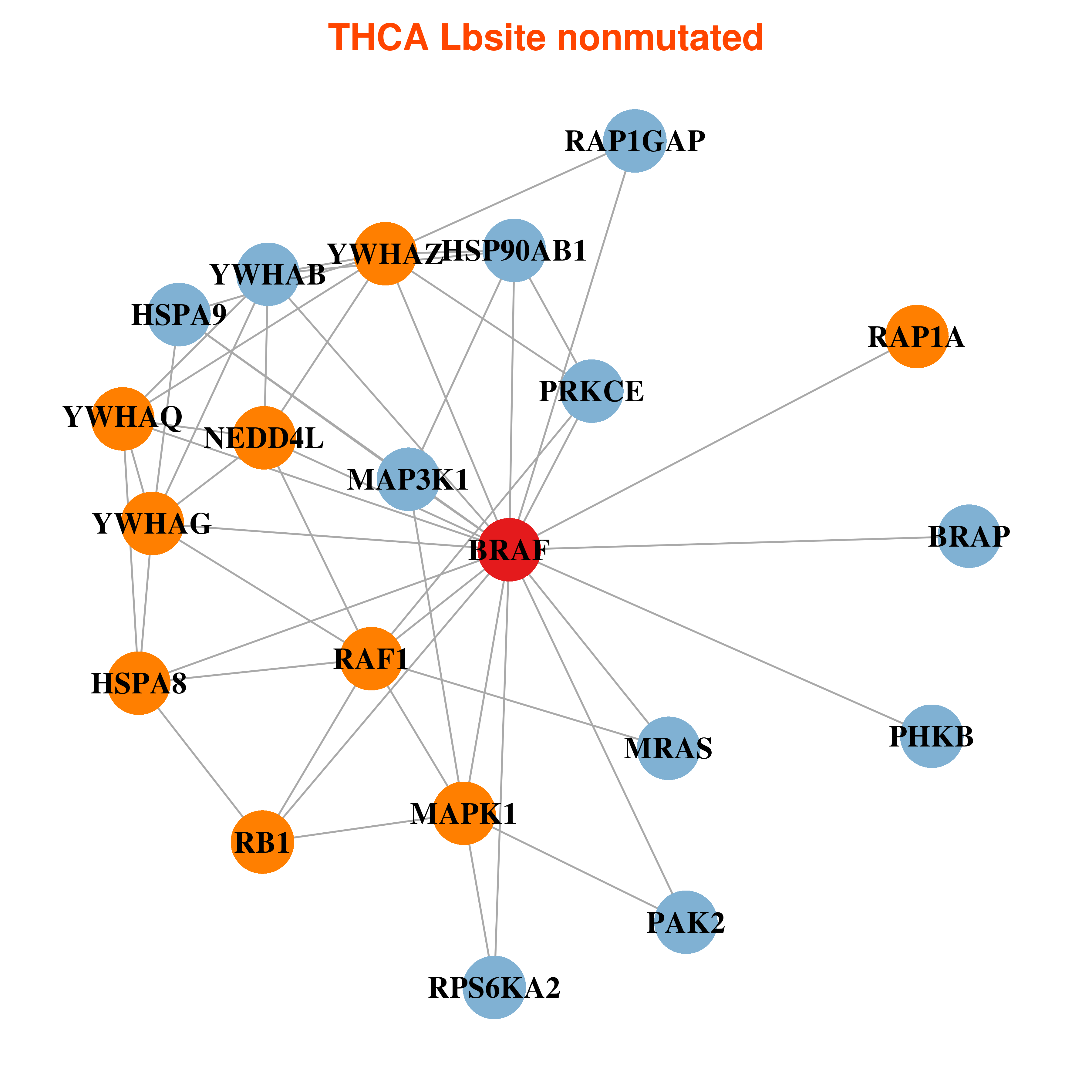
| * In THCA | |
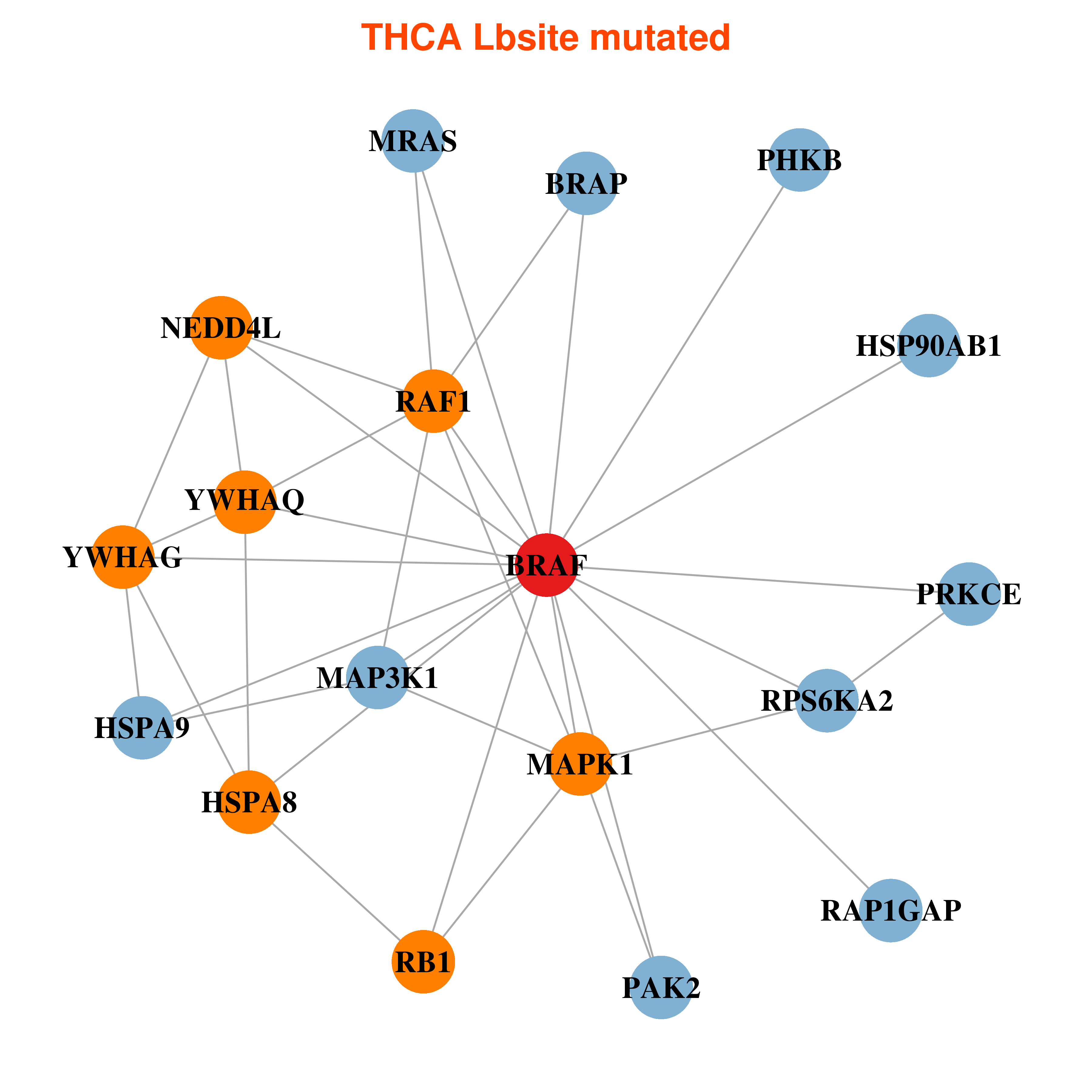 |
|
| Top |
| Top |
| Phenotype information for BRAF |
 Gene level disease information (DisGeNet) Gene level disease information (DisGeNet) |
| Disease ID | Disease name | # PubMed | Association type |
| umls:C0025202 | Melanoma | 688 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C1527249 | Colorectal Cancer | 247 | Biomarker, GeneticVariation |
| umls:C0040136 | Thyroid Neoplasms | 97 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0009404 | Colorectal Neoplasms | 55 | Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0699790 | Colon Carcinoma | 47 | Biomarker, GeneticVariation |
| umls:C1275081 | Cardiofaciocutaneous syndrome | 26 | Biomarker, GeneticVariation |
| umls:C0017638 | Glioma | 23 | Biomarker, GeneticVariation |
| umls:C0007131 | Carcinoma, Non-Small-Cell Lung | 23 | Biomarker, GeneticVariation |
| umls:C0004114 | Astrocytoma | 16 | Biomarker, GeneticVariation |
| umls:C0684249 | Carcinoma of lung | 14 | Biomarker, GeneticVariation |
| umls:C0028326 | Noonan Syndrome | 8 | Biomarker, GeneticVariation |
| umls:C0152013 | Adenocarcinoma of lung | 8 | Biomarker, GeneticVariation |
| umls:C0587248 | Costello Syndrome | 5 | Biomarker |
| umls:C0024121 | Lung Neoplasms | 3 | Biomarker, GeneticVariation |
| umls:C0023903 | Liver Neoplasms | 3 | Biomarker, GeneticVariation |
| umls:C0002448 | Ameloblastoma | 3 | Biomarker, GeneticVariation |
| umls:C0206686 | Adrenocortical Carcinoma | 3 | Biomarker, GeneticVariation |
| umls:C0206754 | Neuroendocrine Tumors | 3 | Biomarker |
| umls:C0024694 | Mandibular Neoplasms | 2 | Biomarker, GeneticVariation |
| umls:C0022665 | Kidney Neoplasms | 2 | Biomarker |
| umls:C0024305 | Lymphoma, Non-Hodgkin | 1 | GeneticVariation |
| umls:C0010276 | Craniopharyngioma | 1 | Biomarker, GeneticVariation |
 Mutation level pathogenic information (ClinVar annotation) Mutation level pathogenic information (ClinVar annotation) |
| Allele ID | AA change | Clinical significance | Origin | Phenotype IDs |
| 29000 | V600E | Pathogenic | Germline;somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0025202 SNOMED CT:2092003 MedGen:C0238463 OMIM:188550 Orphanet:ORPHA146 SNOMED CT:255029007 MedGen:C0699790 SNOMED CT:269533000 MedGen:C1266158 MedGen:CN221809 |
| 29006 | G466V | Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0152013 |
| 29009 | G469R | Pathogenic | Germline;somatic | MedGen:C0024305 OMIM:605027 Orphanet:ORPHA547 SNOMED CT:1929004 MedGen:CN166718 |
| 29010 | G469A | Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0024305 OMIM:605027 Orphanet:ORPHA547 SNOMED CT:1929004 |
| 29011 | D594G | Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 MedGen:C0024305 OMIM:605027 Orphanet:ORPHA547 SNOMED CT:1929004 |
| 29013 | G469E | Pathogenic | Germline;unknown | GeneReviews:NBK1186 MedGen:CN029449 OMIM:115150 Orphanet:ORPHA1340 MedGen:CN166718 |
| 48834 | G464V | Pathogenic | Somatic;unknown | MedGen:C0007131 SNOMED CT:254637007 MedGen:CN166718 |
| 53968 | G466R | Likely pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| 53970 | G469V | Pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| 53980 | D594N | Likely pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| 174177 | N581S | Likely pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| 174179 | G469R | Likely pathogenic | Somatic | MedGen:C0007131 SNOMED CT:254637007 |
| Top |
| Pharmacological information for BRAF |
 Gene expression profile of anticancer drug treated cell-lines (CCLE) Gene expression profile of anticancer drug treated cell-lines (CCLE)Heatmap showing the correlation between gene expression and drug response across all the cell-lines. We chose the top 20 among 138 drugs.We used Pearson's correlation coefficient. |
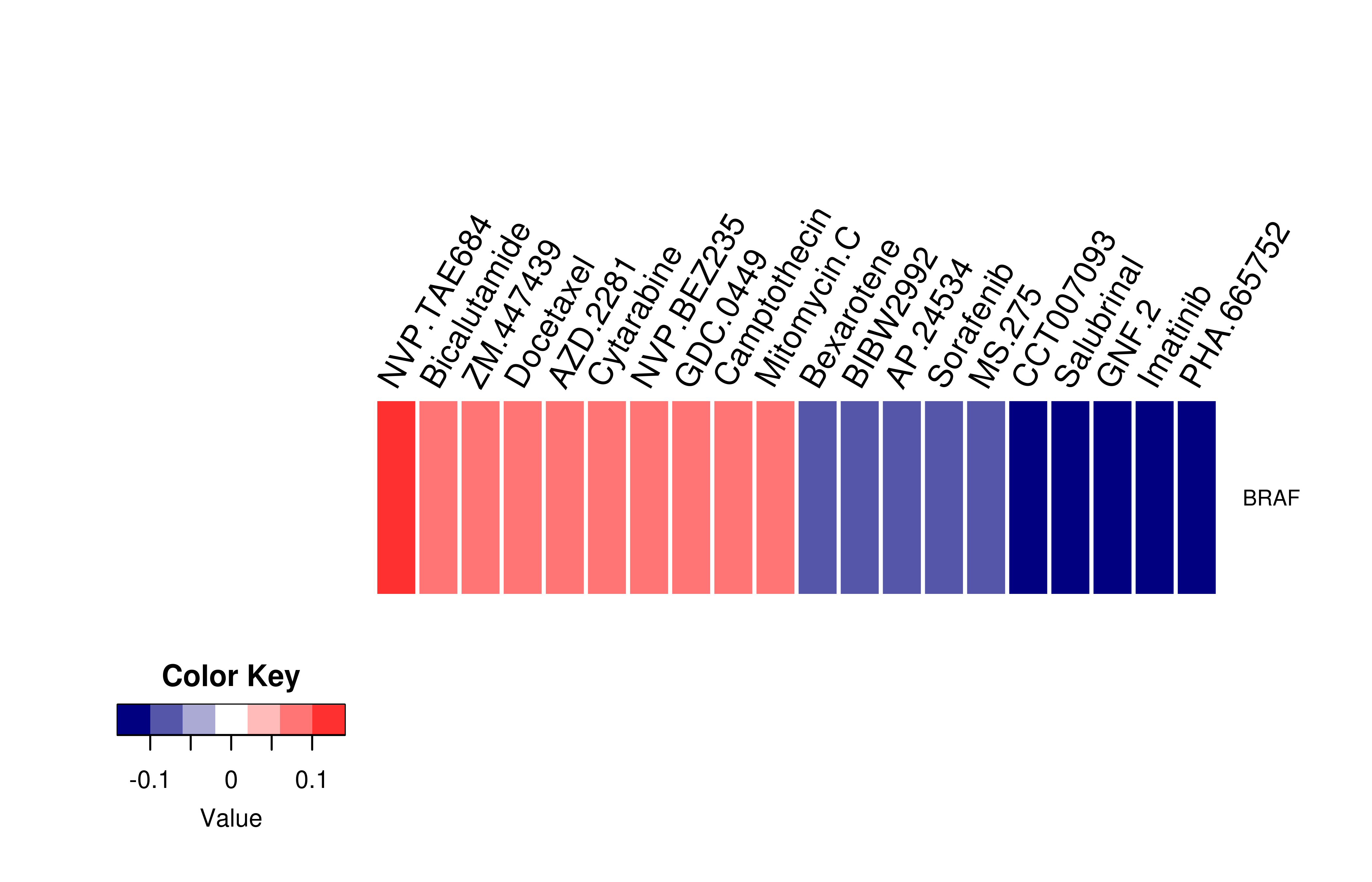 |
 Gene-centered drug-gene interaction network Gene-centered drug-gene interaction network |
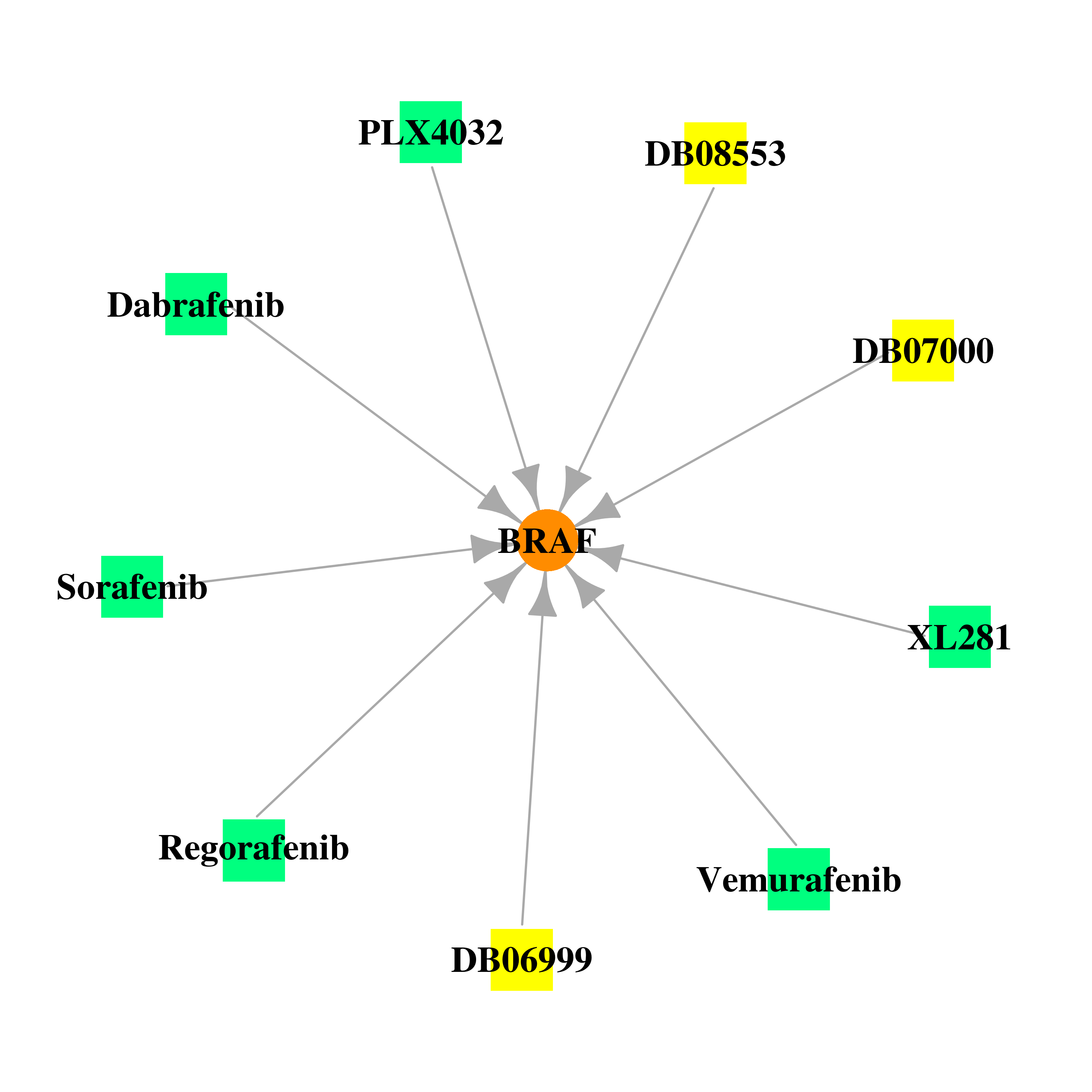 |
 Drug information targeting mutLBSgene (Approved drugs only) Drug information targeting mutLBSgene (Approved drugs only) |
| Drug status | DrugBank ID | Name | Type | Drug structure |
| Approved|investigational | DB00398 | Sorafenib | Small molecule |  |
| Investigational | DB05190 | XL281 | Small molecule |  |
| Investigational | DB05238 | PLX4032 | Small molecule |  |
| Experimental | DB06999 | N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}propane-1-sulfonamide | Small molecule |  |
| Experimental | DB07000 | N-{2,4-difluoro-3-[(5-pyridin-3-yl-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]phenyl}ethanesulfonamide | Small molecule | 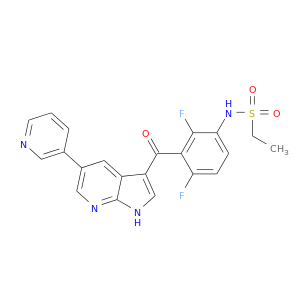 |
| Experimental | DB08553 | (1E)-5-(1-piperidin-4-yl-3-pyridin-4-yl-1H-pyrazol-4-yl)-2,3-dihydro-1H-inden-1-one oxime | Small molecule | 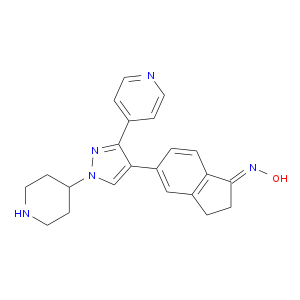 |
| Approved | DB08881 | Vemurafenib | Small molecule |  |
| Approved | DB08896 | Regorafenib | Small molecule | 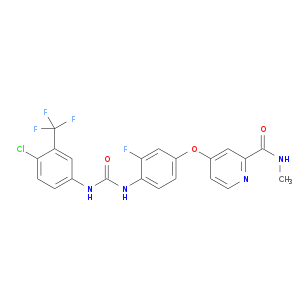 |
| Approved | DB08912 | Dabrafenib | Small molecule | 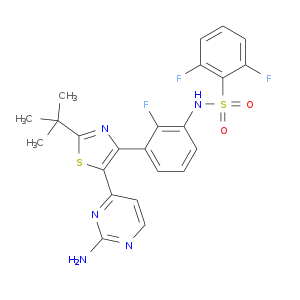 |
 Gene-centered ligand-gene interaction network Gene-centered ligand-gene interaction network |
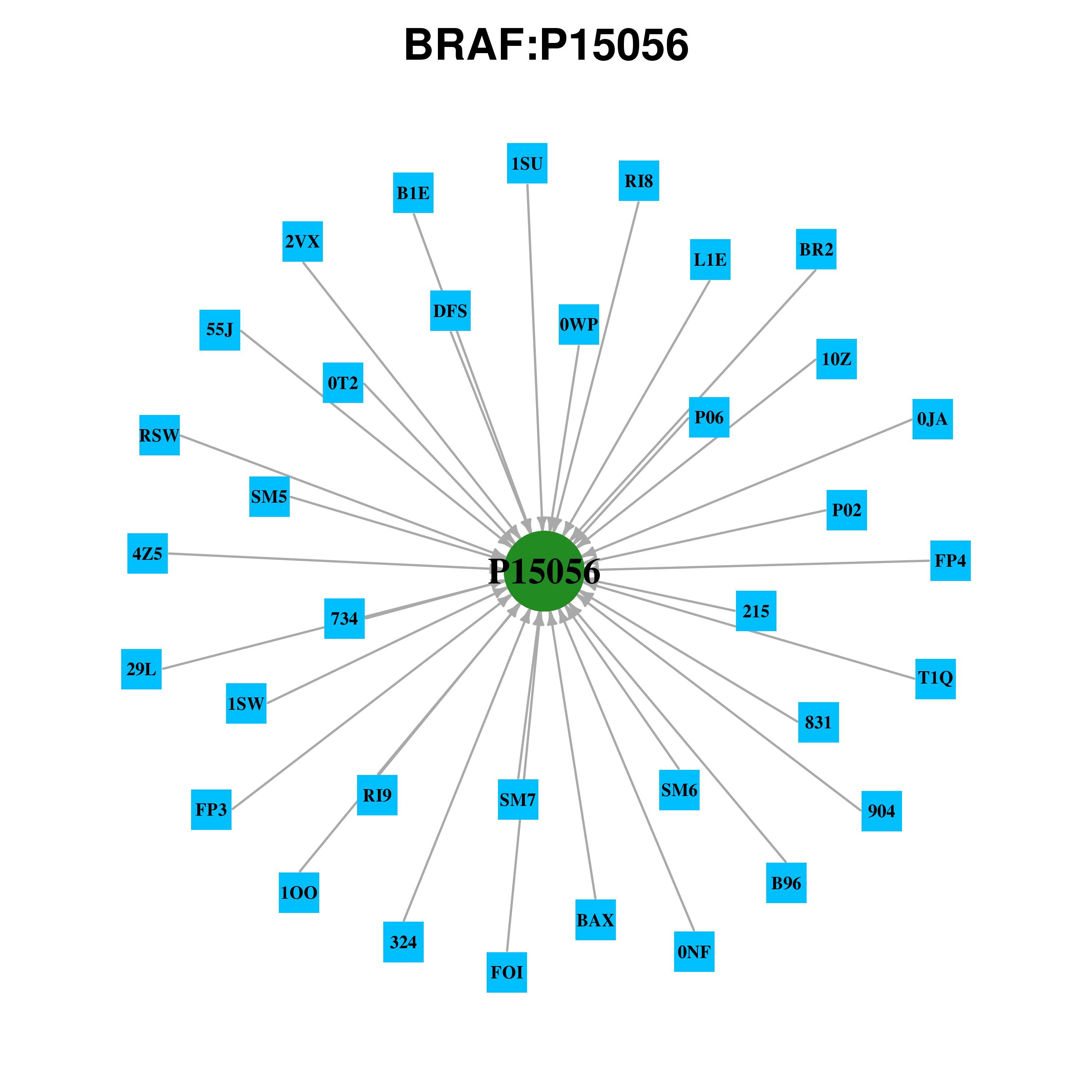 |
 Ligands binding to mutated ligand binding site of BRAF go to BioLip Ligands binding to mutated ligand binding site of BRAF go to BioLip |
| Ligand ID | Ligand short name | Ligand long name | PDB ID | PDB name | mutLBS | P06 | DABRAFENIB | N-{3-[5-(2-AMINOPYRIMIDIN-4-YL)-2-TERT-BUTYL-1,3- THIAZOL-4-YL]-2-FLUOROPHENYL}-2,6- DIFLUOROBENZENESULFONAMIDE | 4xv2 | A | G464 F468 V471 L514 F583 G593 D594 F595 | P06 | DABRAFENIB | N-{3-[5-(2-AMINOPYRIMIDIN-4-YL)-2-TERT-BUTYL-1,3- THIAZOL-4-YL]-2-FLUOROPHENYL}-2,6- DIFLUOROBENZENESULFONAMIDE | 4xv2 | B | G464 G466 F468 V471 L514 F583 G593 D594 F595 | 734 | 5-CHLORO-7-[(R)-FURAN-2-YL(PYRIDIN-2-YLAMINO)METHYL]QUINOLIN-8-OL | 4e26 | B | I463 G464 S536 F583 | 215 | (1Z)-5-(2-{4-[2-(DIMETHYLAMINO)ETHOXY]PHENYL}-5-PYRIDIN-4-YL-1H-IMIDAZOL-4-YL)INDAN-1-ONE OXIME | 2fb8 | A | I463 G464 V471 L514 F583 D594 | SM5 | (1E)-5-(1-PIPERIDIN-4-YL-3-PYRIDIN-4-YL-1H-PYRAZOL-4-YL)-2,3-DIHYDRO-1H-INDEN-1-ONE OXIME | 3d4q | A | I463 G464 V471 L514 F583 D594 | SM5 | (1E)-5-(1-PIPERIDIN-4-YL-3-PYRIDIN-4-YL-1H-PYRAZOL-4-YL)-2,3-DIHYDRO-1H-INDEN-1-ONE OXIME | 3d4q | B | I463 G464 V471 L514 F583 D594 | FP4 | 3-(4-{[2-(PYRIMIDIN-2-YL)FURO[2,3-C]PYRIDIN-3-YL]AMINO}-1H-INDAZOL-3-YL)PROPAN-1-OL | 3pri | A | I463 G466 V471 L514 N581 F583 D594 | FP4 | 3-(4-{[2-(PYRIMIDIN-2-YL)FURO[2,3-C]PYRIDIN-3-YL]AMINO}-1H-INDAZOL-3-YL)PROPAN-1-OL | 3pri | B | I463 G466 V471 N581 F583 D594 | 904 | N'-(3-{[5-(2-CYCLOPROPYLPYRIMIDIN-5-YL)-1H-PYRROLO[2,3- B]PYRIDIN-3-YL]CARBONYL}-2,4-DIFLUOROPHENYL)-N-ETHYL- N-METHYLSULFURIC DIAMIDE | 4xv1 | A | I463 L514 D594 F595 G596 | 0WP | 3-{[3-(2-CYANOPROPAN-2-YL)BENZOYL]AMINO}-2,6-DIFLUORO- N-(3-METHOXY-2H-PYRAZOLO[3,4-B]PYRIDIN-5-YL)BENZAMIDE | 4g9c | A | I463 L514 F583 G593 D594 F595 | 0WP | 3-{[3-(2-CYANOPROPAN-2-YL)BENZOYL]AMINO}-2,6-DIFLUORO- N-(3-METHOXY-2H-PYRAZOLO[3,4-B]PYRIDIN-5-YL)BENZAMIDE | 4g9c | B | I463 L514 F583 G593 D594 F595 | 734 | 5-CHLORO-7-[(R)-FURAN-2-YL(PYRIDIN-2-YLAMINO)METHYL]QUINOLIN-8-OL | 4e26 | A | I463 L514 S536 F583 | 29L | 2-{4-[(1E)-1-(HYDROXYIMINO)-2,3-DIHYDRO-1H-INDEN-5-YL]- 3-(PYRIDIN-4-YL)-1H-PYRAZOL-1-YL}ETHANOL | 4mnf | B | I463 V471 F583 | FOI | METHYL 3-{[(5S)-1-(HYDROXYAMINO)-5H-INDEN-5-YL]AMINO}FURO[2,3-C]PYRIDINE-2-CARBOXYLATE | 3ppj | B | I463 V471 F583 D594 | FNI | 3-[(5-HYDROXYNAPHTHALEN-2-YL)AMINO]-N-(PYRIMIDIN-4-YL)FURO[2,3-C]PYRIDINE-2-CARBOXAMIDE | 3ppk | A | I463 V471 F583 D594 | FNI | 3-[(5-HYDROXYNAPHTHALEN-2-YL)AMINO]-N-(PYRIMIDIN-4-YL)FURO[2,3-C]PYRIDINE-2-CARBOXAMIDE | 3ppk | B | I463 V471 F583 D594 | SM6 | ETHYL 3-{[1-(HYDROXYAMINO)-2H-INDEN-5-YL]AMINO}THIENO[2,3-C]PYRIDINE-2-CARBOXYLATE | 3psb | A | I463 V471 F583 D594 | SM6 | ETHYL 3-{[1-(HYDROXYAMINO)-2H-INDEN-5-YL]AMINO}THIENO[2,3-C]PYRIDINE-2-CARBOXYLATE | 3psb | B | I463 V471 F583 D594 | 2VX | N-{2,4-DIFLUORO-3-[METHYL(3-METHYL-4-OXO-3,4- DIHYDROQUINAZOLIN-6-YL)AMINO]PHENYL}PROPANE-1- SULFONAMIDE | 4pp7 | B | I463 V471 L514 D594 F595 G596 | 324 | N-{3-[(5-CHLORO-1H-PYRROLO[2,3-B]PYRIDIN-3-YL)CARBONYL]-2,4-DIFLUOROPHENYL}PROPANE-1-SULFONAMIDE | 3c4c | B | I463 V471 L514 F583 D594 | FOI | METHYL 3-{[(5S)-1-(HYDROXYAMINO)-5H-INDEN-5-YL]AMINO}FURO[2,3-C]PYRIDINE-2-CARBOXYLATE | 3ppj | A | I463 V471 L514 F583 D594 | FP3 | 2-CHLORO-5-{[2-(PYRIMIDIN-2-YL)FURO[2,3-C]PYRIDIN-3-YL]AMINO}PHENOL | 3prf | B | I463 V471 L514 F583 D594 | SM7 | 6-[1-(PIPERIDIN-4-YL)-3-(PYRIDIN-4-YL)-1H-PYRAZOL-4-YL]INDENO[1,2-C]PYRAZOLE | 3psd | A | I463 V471 L514 F583 D594 | SM7 | 6-[1-(PIPERIDIN-4-YL)-3-(PYRIDIN-4-YL)-1H-PYRAZOL-4-YL]INDENO[1,2-C]PYRAZOLE | 3psd | B | I463 V471 L514 F583 D594 | 324 | N-{3-[(5-CHLORO-1H-PYRROLO[2,3-B]PYRIDIN-3-YL)CARBONYL]-2,4-DIFLUOROPHENYL}PROPANE-1-SULFONAMIDE | 3c4c | A | I463 V471 L514 F583 D594 F595 | 325 | N-{2,4-DIFLUORO-3-[(5-PYRIDIN-3-YL-1H-PYRROLO[2,3-B]PYRIDIN-3-YL)CARBONYL]PHENYL}ETHANESULFONAMIDE | 4fk3 | A | I463 V471 L514 F583 D594 F595 G596 | FP3 | 2-CHLORO-5-{[2-(PYRIMIDIN-2-YL)FURO[2,3-C]PYRIDIN-3-YL]AMINO}PHENOL | 3prf | A | I463 V471 L514 F583 G593 D594 | 55J | 1-METHYL-5-({2-[5-(TRIFLUOROMETHYL)-1H-IMIDAZOL-2- YL]PYRIDIN-4-YL}OXY)-N-[4-(TRIFLUOROMETHYL)PHENYL]-1H- BENZIMIDAZOL-2-AMINE | 5ct7 | A | I463 V471 L514 F583 I592 G593 D594 F595 | 55J | 1-METHYL-5-({2-[5-(TRIFLUOROMETHYL)-1H-IMIDAZOL-2- YL]PYRIDIN-4-YL}OXY)-N-[4-(TRIFLUOROMETHYL)PHENYL]-1H- BENZIMIDAZOL-2-AMINE | 5ct7 | B | I463 V471 L514 F583 I592 G593 D594 F595 | 1OO | N-{3-[(5-CHLORO-1H-PYRROLO[2,3-B]PYRIDIN-3-YL) CARBONYL]-2,4-DIFLUOROPHENYL}-4-(TRIFLUOROMETHYL) BENZENESULFONAMIDE | 4xv9 | A | I463 V471 L514 F583 I592 G593 D594 F595 | B1E | 3-(2-CYANOPROPAN-2-YL)-N-{4-METHYL-3-[(3-METHYL-4-OXO- 3,4-DIHYDROQUINAZOLIN-6-YL)AMINO]PHENYL}BENZAMIDE | 4g9r | B | I463 V471 L514 G593 D594 F595 | 215 | (1Z)-5-(2-{4-[2-(DIMETHYLAMINO)ETHOXY]PHENYL}-5-PYRIDIN-4-YL-1H-IMIDAZOL-4-YL)INDAN-1-ONE OXIME | 2fb8 | B | I463 V471 Y538 F583 D594 | BR2 | 2,6-DIFLUORO-N-[(5S)-3-METHOXY-5H-PYRAZOLO[3,4-B]PYRIDIN-5-YL]-3-[(PHENYLSULFONYL)AMINO]BENZAMIDE | 3skc | B | L514 F583 D594 F595 G596 | TV4 | N-(6-AMINO-5-BROMOPYRIDIN-3-YL)-2,6-DIFLUORO-3-[(PROPYLSULFONYL)AMINO]BENZAMIDE | 3tv4 | A | L514 F583 D594 F595 G596 | B0R | 2,6-DIFLUORO-N-(3-METHOXY-2H-PYRAZOLO[3,4-B]PYRIDIN-5-YL)-3-[(PROPYLSULFONYL)AMINO]BENZAMIDE | 3tv6 | A | L514 F583 D594 F595 G596 | RI9 | N-{2,4-DIFLUORO-3-[({6-[(2-HYDROXYETHYL) AMINO]PYRIMIDIN-4-YL}CARBAMOYL)AMINO]PHENYL}PROPANE-1- SULFONAMIDE | 4ehg | B | L514 F583 D594 F595 G596 | B0R | 2,6-DIFLUORO-N-(3-METHOXY-2H-PYRAZOLO[3,4-B]PYRIDIN-5-YL)-3-[(PROPYLSULFONYL)AMINO]BENZAMIDE | 3tv6 | B | L514 F583 D594 F595 G596 L597 | B96 | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA | 4jvg | A | L514 F583 G593 D594 F595 | B96 | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA | 4jvg | D | L514 F583 G593 D594 F595 | 0NF | (2S)-N-[3-(2-AMINOPROPAN-2-YL)-5-(TRIFLUOROMETHYL)PHENYL]-7-[(7-OXO-5,6,7,8-TETRAHYDRO-1,8-NAPHTHYRIDIN-4-YL)OXY]-1,2,3,4-TETRAHYDRONAPHTHALENE-2-CARBOXAMIDE | 3q96 | A | L514 G593 D594 F595 | BR2 | 2,6-DIFLUORO-N-[(5S)-3-METHOXY-5H-PYRAZOLO[3,4-B]PYRIDIN-5-YL]-3-[(PHENYLSULFONYL)AMINO]BENZAMIDE | 3skc | A | L514 G593 D594 F595 | B1E | 3-(2-CYANOPROPAN-2-YL)-N-{4-METHYL-3-[(3-METHYL-4-OXO- 3,4-DIHYDROQUINAZOLIN-6-YL)AMINO]PHENYL}BENZAMIDE | 4g9r | A | L514 G593 D594 F595 | DFS | 2,6-DIFLUORO-N-(1H-IMIDAZO[4,5-B]PYRIDIN-6-YL)-3- [(PROPYLSULFONYL)AMINO]BENZAMIDE | 4mbj | B | L514 G593 F595 G596 | T1Q | N-(2,4-DIFLUORO-3-{2-[(3-HYDROXYPROPYL)AMINO]-8-METHYL-7-OXO-7,8-DIHYDROPYRIDO[2,3-D]PYRIMIDIN-6-YL}PHENYL)PROPANE-1-SULFONAMIDE | 4e4x | A | L514 S536 F583 D594 F595 G596 | 29L | 2-{4-[(1E)-1-(HYDROXYIMINO)-2,3-DIHYDRO-1H-INDEN-5-YL]- 3-(PYRIDIN-4-YL)-1H-PYRAZOL-1-YL}ETHANOL | 4mnf | A | V471 L514 F583 D594 | TV4 | N-(6-AMINO-5-BROMOPYRIDIN-3-YL)-2,6-DIFLUORO-3-[(PROPYLSULFONYL)AMINO]BENZAMIDE | 3tv4 | B | V471 L514 F583 D594 F595 G596 | RI8 | 4-AMINO-N-{2,6-DIFLUORO-3-[(PROPYLSULFONYL) AMINO]PHENYL}THIENO[3,2-D]PYRIMIDINE-7-CARBOXAMIDE | 4ehe | A | V471 L514 F583 D594 F595 G596 | RI8 | 4-AMINO-N-{2,6-DIFLUORO-3-[(PROPYLSULFONYL) AMINO]PHENYL}THIENO[3,2-D]PYRIMIDINE-7-CARBOXAMIDE | 4ehe | B | V471 L514 F583 D594 F595 G596 | RI9 | N-{2,4-DIFLUORO-3-[({6-[(2-HYDROXYETHYL) AMINO]PYRIMIDIN-4-YL}CARBAMOYL)AMINO]PHENYL}PROPANE-1- SULFONAMIDE | 4ehg | A | V471 L514 F583 D594 F595 G596 | 2VX | N-{2,4-DIFLUORO-3-[METHYL(3-METHYL-4-OXO-3,4- DIHYDROQUINAZOLIN-6-YL)AMINO]PHENYL}PROPANE-1- SULFONAMIDE | 4pp7 | A | V471 L514 F583 D594 F595 G596 | P02 | N'-{3-[5-(2-AMINOPYRIMIDIN-4-YL)-2-TERT-BUTYL-1,3- THIAZOL-4-YL]-2-FLUOROPHENYL}-N-ETHYL-N-METHYLSULFURIC DIAMIDE | 4xv3 | A | V471 L514 F583 D594 F595 G596 | L1E | N5-(3-(9H-PURIN-6-YL)PYRIDIN-2-YL)-N1-(4-CHLOROPHENYL)-6-METHYLISOQUINOLINE-1,5-DIAMINE | 3idp | B | V471 L514 F583 G593 D594 F595 | 0NF | (2S)-N-[3-(2-AMINOPROPAN-2-YL)-5-(TRIFLUOROMETHYL)PHENYL]-7-[(7-OXO-5,6,7,8-TETRAHYDRO-1,8-NAPHTHYRIDIN-4-YL)OXY]-1,2,3,4-TETRAHYDRONAPHTHALENE-2-CARBOXAMIDE | 3q96 | B | V471 L514 F583 G593 D594 F595 A598 | L1E | N5-(3-(9H-PURIN-6-YL)PYRIDIN-2-YL)-N1-(4-CHLOROPHENYL)-6-METHYLISOQUINOLINE-1,5-DIAMINE | 3idp | A | V471 L514 G593 D594 F595 | B96 | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA | 4jvg | B | V471 L514 G593 D594 F595 | B96 | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA | 4jvg | C | V471 L514 G593 D594 F595 | DFS | 2,6-DIFLUORO-N-(1H-IMIDAZO[4,5-B]PYRIDIN-6-YL)-3- [(PROPYLSULFONYL)AMINO]BENZAMIDE | 4mbj | A | V471 L514 G593 D594 F595 G596 L597 | T1Q | N-(2,4-DIFLUORO-3-{2-[(3-HYDROXYPROPYL)AMINO]-8-METHYL-7-OXO-7,8-DIHYDROPYRIDO[2,3-D]PYRIMIDIN-6-YL}PHENYL)PROPANE-1-SULFONAMIDE | 4e4x | B | V471 L514 S536 F583 D594 F595 G596 |
| Top |
| Conservation information for LBS of BRAF |
 Multiple alignments for P15056 in multiple species Multiple alignments for P15056 in multiple species |
| LBS | AA sequence | # species | Species | A481 | WHGDVAVKMLN | 3 | Homo sapiens, Gallus gallus, Mus musculus | A481 | FFGTVAIKKLN | 1 | Caenorhabditis elegans | A481 | WHGPVAVKTLN | 1 | Drosophila melanogaster | A598 | GDFGLATVKSR | 3 | Homo sapiens, Gallus gallus, Mus musculus | A598 | GDFGLATVKTK | 1 | Caenorhabditis elegans | A598 | GDFGLATAKTR | 1 | Drosophila melanogaster | C532 | IVTQWCEGSSL | 4 | Homo sapiens, Drosophila melanogaster, Gallus gallus, Mus musculus | C532 | IITQWCEGSSL | 1 | Caenorhabditis elegans | D594 | TVKIGDFGLAT | 4 | Homo sapiens, Caenorhabditis elegans, Gallus gallus, Mus musculus | D594 | SVKIGDFGLAT | 1 | Drosophila melanogaster | E501 | QAFKNEVGVLR | 3 | Homo sapiens, Gallus gallus, Mus musculus | E501 | AAFKNEVAVLK | 1 | Caenorhabditis elegans | E501 | QAFKNEVAMLK | 1 | Drosophila melanogaster | E533 | VTQWCEGSSLY | 4 | Homo sapiens, Drosophila melanogaster, Gallus gallus, Mus musculus | E533 | ITQWCEGSSLY | 1 | Caenorhabditis elegans | F468 | IGSGSFGTVYK | 3 | Homo sapiens, Gallus gallus, Mus musculus | F468 | VGSGSFGTVYR | 1 | Caenorhabditis elegans | F468 | IGSGSFGTVYR | 1 | Drosophila melanogaster | F516 | VNILLFMGYST | 3 | Homo sapiens, Gallus gallus, Mus musculus | F516 | LNVLLFMGWVR | 1 | Caenorhabditis elegans | F516 | CNILLFMGCVS | 1 | Drosophila melanogaster | F583 | KSNNIFLHEDL | 4 | Homo sapiens, Drosophila melanogaster, Gallus gallus, Mus musculus | F583 | KTNNIFLMDDM | 1 | Caenorhabditis elegans | F595 | VKIGDFGLATV | 4 | Homo sapiens, Caenorhabditis elegans, Gallus gallus, Mus musculus | F595 | VKIGDFGLATA | 1 | Drosophila melanogaster | G464 | VGQRIGSGSFG | 3 | Homo sapiens, Gallus gallus, Mus musculus | G464 | IQYKVGSGSFG | 1 | Caenorhabditis elegans | G464 | IGPRIGSGSFG | 1 | Drosophila melanogaster | G466 | QRIGSGSFGTV | 3 | Homo sapiens, Gallus gallus, Mus musculus | G466 | YKVGSGSFGTV | 1 | Caenorhabditis elegans | G466 | PRIGSGSFGTV | 1 | Drosophila melanogaster | G534 | TQWCEGSSLYH | 3 | Homo sapiens, Gallus gallus, Mus musculus | G534 | TQWCEGSSLYR | 1 | Caenorhabditis elegans | G534 | TQWCEGSSLYK | 1 | Drosophila melanogaster | G593 | -TVKIGDFGLA | 3 | Homo sapiens, Gallus gallus, Mus musculus | G593 | STVKIGDFGLA | 1 | Caenorhabditis elegans | G593 | -SVKIGDFGLA | 1 | Drosophila melanogaster | G596 | KIGDFGLATVK | 4 | Homo sapiens, Caenorhabditis elegans, Gallus gallus, Mus musculus | G596 | KIGDFGLATAK | 1 | Drosophila melanogaster | H539 | GSSLYHHLHII | 3 | Homo sapiens, Gallus gallus, Mus musculus | H539 | GSSLYRHIHVQ | 1 | Caenorhabditis elegans | H539 | GSSLYKHVHVS | 1 | Drosophila melanogaster | H574 | AKSIIHRDLKS | 3 | Homo sapiens, Gallus gallus, Mus musculus | H574 | SKNIIHRDLKT | 1 | Caenorhabditis elegans | H574 | AKNIIHRDLKS | 1 | Drosophila melanogaster | I463 | TVGQRIGSGSF | 3 | Homo sapiens, Gallus gallus, Mus musculus | I463 | IIQYKVGSGSF | 1 | Caenorhabditis elegans | I463 | LIGPRIGSGSF | 1 | Drosophila melanogaster | I513 | TRHVNILLFMG | 3 | Homo sapiens, Gallus gallus, Mus musculus | I513 | TRHLNVLLFMG | 1 | Caenorhabditis elegans | I513 | TRHCNILLFMG | 1 | Drosophila melanogaster | I527 | KPQLAIVTQWC | 3 | Homo sapiens, Gallus gallus, Mus musculus | I527 | EPEIAIITQWC | 1 | Caenorhabditis elegans | I527 | KPSLAIVTQWC | 1 | Drosophila melanogaster | I543 | YHHLHIIETK- | 3 | Homo sapiens, Gallus gallus, Mus musculus | I543 | YRHIHVQEPRV | 1 | Caenorhabditis elegans | I543 | YKHVHVSETK- | 1 | Drosophila melanogaster | I572 | LHAKSIIHRDL | 3 | Homo sapiens, Gallus gallus, Mus musculus | I572 | LHSKNIIHRDL | 1 | Caenorhabditis elegans | I572 | LHAKNIIHRDL | 1 | Drosophila melanogaster | I592 | L-TVKIGDFGL | 3 | Homo sapiens, Gallus gallus, Mus musculus | I592 | MSTVKIGDFGL | 1 | Caenorhabditis elegans | I592 | L-SVKIGDFGL | 1 | Drosophila melanogaster | K483 | GDVAVKMLNVT | 3 | Homo sapiens, Gallus gallus, Mus musculus | K483 | GTVAIKKLNVV | 1 | Caenorhabditis elegans | K483 | GPVAVKTLNVK | 1 | Drosophila melanogaster | L505 | NEVGVLRKTRH | 3 | Homo sapiens, Gallus gallus, Mus musculus | L505 | NEVAVLKKTRH | 1 | Caenorhabditis elegans | L505 | NEVAMLKKTRH | 1 | Drosophila melanogaster | L514 | RHVNILLFMGY | 3 | Homo sapiens, Gallus gallus, Mus musculus | L514 | RHLNVLLFMGW | 1 | Caenorhabditis elegans | L514 | RHCNILLFMGC | 1 | Drosophila melanogaster | L567 | QGMDYLHAKSI | 3 | Homo sapiens, Gallus gallus, Mus musculus | L567 | LGMNYLHSKNI | 1 | Caenorhabditis elegans | L567 | QGMDYLHAKNI | 1 | Drosophila melanogaster | L597 | IGDFGLATVKS | 3 | Homo sapiens, Gallus gallus, Mus musculus | L597 | IGDFGLATVKT | 1 | Caenorhabditis elegans | L597 | IGDFGLATAKT | 1 | Drosophila melanogaster | N580 | RDLKSNNIFLH | 4 | Homo sapiens, Drosophila melanogaster, Gallus gallus, Mus musculus | N580 | RDLKTNNIFLM | 1 | Caenorhabditis elegans | N581 | DLKSNNIFLHE | 4 | Homo sapiens, Drosophila melanogaster, Gallus gallus, Mus musculus | N581 | DLKTNNIFLMD | 1 | Caenorhabditis elegans | Q530 | LAIVTQWCEGS | 4 | Homo sapiens, Drosophila melanogaster, Gallus gallus, Mus musculus | Q530 | IAIITQWCEGS | 1 | Caenorhabditis elegans | S465 | GQRIGSGSFGT | 3 | Homo sapiens, Gallus gallus, Mus musculus | S465 | QYKVGSGSFGT | 1 | Caenorhabditis elegans | S465 | GPRIGSGSFGT | 1 | Drosophila melanogaster | S535 | QWCEGSSLYHH | 3 | Homo sapiens, Gallus gallus, Mus musculus | S535 | QWCEGSSLYRH | 1 | Caenorhabditis elegans | S535 | QWCEGSSLYKH | 1 | Drosophila melanogaster | S536 | WCEGSSLYHHL | 3 | Homo sapiens, Gallus gallus, Mus musculus | S536 | WCEGSSLYRHI | 1 | Caenorhabditis elegans | S536 | WCEGSSLYKHV | 1 | Drosophila melanogaster | T508 | GVLRKTRHVNI | 3 | Homo sapiens, Gallus gallus, Mus musculus | T508 | AVLKKTRHLNV | 1 | Caenorhabditis elegans | T508 | AMLKKTRHCNI | 1 | Drosophila melanogaster | T529 | QLAIVTQWCEG | 3 | Homo sapiens, Gallus gallus, Mus musculus | T529 | EIAIITQWCEG | 1 | Caenorhabditis elegans | T529 | SLAIVTQWCEG | 1 | Drosophila melanogaster | V471 | GSFGTVYKGKW | 3 | Homo sapiens, Gallus gallus, Mus musculus | V471 | GSFGTVYRGEF | 1 | Caenorhabditis elegans | V471 | GSFGTVYRAHW | 1 | Drosophila melanogaster | V482 | HGDVAVKMLNV | 3 | Homo sapiens, Gallus gallus, Mus musculus | V482 | FGTVAIKKLNV | 1 | Caenorhabditis elegans | V482 | HGPVAVKTLNV | 1 | Drosophila melanogaster | V504 | KNEVGVLRKTR | 3 | Homo sapiens, Gallus gallus, Mus musculus | V504 | KNEVAVLKKTR | 1 | Caenorhabditis elegans | V504 | KNEVAMLKKTR | 1 | Drosophila melanogaster | W531 | AIVTQWCEGSS | 4 | Homo sapiens, Drosophila melanogaster, Gallus gallus, Mus musculus | W531 | AIITQWCEGSS | 1 | Caenorhabditis elegans | Y538 | EGSSLYHHLHI | 3 | Homo sapiens, Gallus gallus, Mus musculus | Y538 | EGSSLYRHIHV | 1 | Caenorhabditis elegans | Y538 | EGSSLYKHVHV | 1 | Drosophila melanogaster |
 |
Copyright © 2016-Present - The University of Texas Health Science Center at Houston |