|
mutLBSgeneDB |
| |
| |
| |
| |
| |
| |
|
| Gene summary for TP53 |
 Gene summary Gene summary |
| Basic gene Info. | Gene symbol | TP53 |
| Gene name | tumor protein p53 | |
| Synonyms | BCC7|LFS1|P53|TRP53 | |
| Cytomap | UCSC genome browser: 17p13.1 | |
| Type of gene | protein-coding | |
| RefGenes | NM_000546.5, NM_001126112.2,NM_001126113.2,NM_001126114.2,NM_001126115.1, NM_001126116.1,NM_001126117.1,NM_001126118.1,NM_001276695.1, NM_001276696.1,NM_001276697.1,NM_001276698.1,NM_001276699.1, NM_001276760.1,NM_001276761.1, | |
| Description | antigen NY-CO-13cellular tumor antigen p53mutant tumor protein 53p53 tumor suppressorphosphoprotein p53transformation-related protein 53tumor protein 53 | |
| Modification date | 20141222 | |
| dbXrefs | MIM : 191170 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000141510 | ||
| HPRD : 01859 | ||
| Vega : OTTHUMG00000162125 | ||
| Protein | UniProt: P04637 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_TP53 | |
| BioGPS: 7157 | ||
| Pathway | NCI Pathway Interaction Database: TP53 | |
| KEGG: TP53 | ||
| REACTOME: TP53 | ||
| Pathway Commons: TP53 | ||
| Context | iHOP: TP53 | |
| ligand binding site mutation search in PubMed: TP53 | ||
| UCL Cancer Institute: TP53 | ||
| Assigned class in mutLBSgeneDB | A: This gene has a literature evidence and it belongs to targetable_mutLBSgenes. | |
| References showing study about ligand binding site mutation for TP53. | 1. "Archibald KM, Kulbe H, Kwong J, Chakravarty P, Temple J, Chaplin T, Flak MB, McNeish IA, Deen S, Brenton JD, Young BD, Balkwill F. Sequential genetic change at the TP53 and chemokine receptor CXCR4 locus during transformation of human ovarian surface epithelium. Oncogene. 2012 Nov 29;31(48):4987-95. doi:10.1038/onc.2011.653. Epub 2012 Jan 23." 22266861 | |
 Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez |
| GO ID | GO Term | PubMed ID | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 15340061 | GO:0000733 | DNA strand renaturation | 8183576 | GO:0006355 | regulation of transcription, DNA-templated | 7587074 | GO:0006461 | protein complex assembly | 12915590 | GO:0006974 | cellular response to DNA damage stimulus | 14744935 | GO:0006983 | ER overload response | 14744935 | GO:0007050 | cell cycle arrest | 15149599 | GO:0008104 | protein localization | 16507995 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 15149599 | GO:0031497 | chromatin assembly | 16322561 | GO:0032461 | positive regulation of protein oligomerization | 12667443 | GO:0034613 | cellular protein localization | 15340061 | GO:0042149 | cellular response to glucose starvation | 14744935 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 14654789 | GO:0042981 | regulation of apoptotic process | 14744935 | GO:0043065 | positive regulation of apoptotic process | 12667443 | GO:0043066 | negative regulation of apoptotic process | 16131611 | GO:0045893 | positive regulation of transcription, DNA-templated | 16322561 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 11672523 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 12667443 | GO:0097252 | oligodendrocyte apoptotic process | 7720704 |
| Top |
| Ligand binding site mutations for TP53 |
 Cancer type specific mutLBS sorted by frequency Cancer type specific mutLBS sorted by frequency |
| LBS | AAchange of nsSNV | Cancer type | # samples | C176 | R175H | BRCA | 15 | C176 | R175H | COAD | 15 | R273 | R273H | OV | 10 | R248 | R248W | COAD | 8 | R248 | R248Q | OV | 8 | E221 | Y220C | OV | 8 | C176 | R175H | OV | 8 | R273 | R273H | COAD | 7 | R283 | R282W | HNSC | 7 | C176 | R175H | HNSC | 7 | R273 | R273H | HNSC | 7 | R273 | R273C | OV | 7 | E221 | Y220C | BRCA | 6 | R248 | R248W | BRCA | 6 | R248 | R248Q | GBM | 6 | R248 | R248W | HNSC | 6 | R273 | R273H | STAD | 6 | R273 | R273C | BRCA | 5 | C176 | R175H | GBM | 5 | M243 | G245S | HNSC | 5 | R248 | R248Q | HNSC | 5 | V157 | R158L | LUSC | 5 | C176 | C176Y | OV | 5 | R283 | R282W | OV | 5 | V157 | V157F | OV | 5 | R273 | R273C | STAD | 5 | C176 | R175H | STAD | 5 | R248 | R248W | UCEC | 5 | C176 | C176F | BRCA | 4 | R273 | R273H | BRCA | 4 | M243 | G245S | COAD | 4 | R273 | R273C | COAD | 4 | R283 | R282W | GBM | 4 | R273 | R273H | GBM | 4 | V157 | A159P | LUAD | 4 | V157 | R158L | LUAD | 4 | S241 | S241F | OV | 4 | R248 | R248W | OV | 4 | R248 | R248Q | STAD | 4 | R283 | R282W | STAD | 4 | R273 | R273C | UCEC | 4 | R248 | R248Q | UCEC | 4 | R273 | R273H | UCEC | 4 | V157 | A159V | BLCA | 3 | K139 | C141Y | BRCA | 3 | M243 | G245D | BRCA | 3 | C238 | M237T | COAD | 3 | R283 | R282W | COAD | 3 | D228 | S227P | COAD | 3 | C275 | C275Y | COAD | 3 | V157 | R158H | GBM | 3 | M243 | G245S | GBM | 3 | E221 | Y220C | GBM | 3 | R248 | R248W | GBM | 3 | V157 | V157F | HNSC | 3 | G279,C277 | P278S | HNSC | 3 | C176 | R175H | LUAD | 3 | R248 | R249M | LUAD | 3 | C238 | M237I | LUAD | 3 | C242 | C242F | LUSC | 3 | R273 | R273L | LUSC | 3 | Q165 | Y163C | LUSC | 3 | V157 | V157F | LUSC | 3 | C176 | R175G | LUSC | 3 | M243 | G245S | OV | 3 | C275 | C275Y | OV | 3 | S241 | S241F | SKCM | 3 | E221 | Y220C | UCEC | 3 | C238 | C238Y | BRCA | 2 | Q165 | Y163C | BRCA | 2 | R283 | R282W | BRCA | 2 | E221 | Y220S | BRCA | 2 | R248 | R248Q | BRCA | 2 | C238 | C238F | BRCA | 2 | F109 | F109L | COAD | 2 | T123 | T125M | COAD | 2 | F109 | F109C | COAD | 2 | M243 | G244D | COAD | 2 | V157 | A159V | COAD | 2 | M243 | G244S | COAD | 2 | V157 | R158H | COAD | 2 | C176 | R175C | COAD | 2 | C238 | Y236C | COAD | 2 | V157 | A159P | COAD | 2 | T155 | T155N | GBM | 2 | C238 | C238F | GBM | 2 | C238 | C238Y | GBM | 2 | L145 | V143M | HNSC | 2 | C238 | Y236C | HNSC | 2 | C238 | C238F | HNSC | 2 | C242 | C242F | HNSC | 2 | C275 | C275F | HNSC | 2 | R273 | R273C | HNSC | 2 | M243 | G245V | HNSC | 2 | R248 | R249S | HNSC | 2 | F109 | R110L | HNSC | 2 | E221 | Y220C | HNSC | 2 | R283 | E285K | HNSC | 2 | C277 | C277F | LUAD | 2 | R280 | D281N | LUAD | 2 | R248 | R249G | LUAD | 2 | F109 | R110L | LUAD | 2 | C275 | C275F | LUAD | 2 | R273 | R273L | LUAD | 2 | L257 | E258D | LUAD | 2 | M243 | G244C | LUAD | 2 | R273 | E271K | LUSC | 2 | E221 | Y220C | LUSC | 2 | M243 | G245V | LUSC | 2 | C238 | M237I | OV | 2 | L257 | D259Y | OV | 2 | Q165 | Y163C | OV | 2 | V157 | A159V | OV | 2 | R273 | R273L | OV | 2 | R273 | V272M | OV | 2 | M243 | G245D | OV | 2 | G279,C277 | P278R | OV | 2 | C176 | P177R | OV | 2 | C238 | M237K | OV | 2 | G279,C277 | P278L | OV | 2 | R248 | P250L | OV | 2 | M243 | G245V | OV | 2 | M243 | G244C | OV | 2 | C238 | Y236C | OV | 2 | M243 | G244S | OV | 2 | E221 | Y220C | STAD | 2 | C176 | C176F | STAD | 2 | V157 | R158C | UCEC | 2 | S241,N239 | S240G | UCEC | 2 | C176 | R175H | UCEC | 2 | M243 | G244S | UCEC | 2 | E221 | Y220C | BLCA | 1 | W146 | W146S | BLCA | 1 | G279 | G279R | BLCA | 1 | K139 | K139N | BLCA | 1 | R273 | R273C | BLCA | 1 | C277 | C277F | BLCA | 1 | C275 | C275Y | BLCA | 1 | C238 | C238F | BLCA | 1 | R273 | R273S | BLCA | 1 | C242 | C242F | BLCA | 1 | R283 | E285K | BLCA | 1 | D148 | D148N | BLCA | 1 | P151 | P151H | BLCA | 1 | V157 | A159P | BLCA | 1 | V157 | R158H | BLCA | 1 | F109 | Y107C | BRCA | 1 | R273 | V272M | BRCA | 1 | G279,C277 | P278L | BRCA | 1 | L257 | I255S | BRCA | 1 | T155 | T155P | BRCA | 1 | E221 | Y220H | BRCA | 1 | G279 | G279E | BRCA | 1 | R248 | M246I | BRCA | 1 | F109 | L111P | BRCA | 1 | R273 | R273P | BRCA | 1 | R280 | D281V | BRCA | 1 | G279,C277 | P278T | BRCA | 1 | M243 | G245S | BRCA | 1 | P151 | P151H | BRCA | 1 | R273,C275 | V274F | BRCA | 1 | V157 | R158G | BRCA | 1 | L257 | I255N | BRCA | 1 | L257 | E258Q | BRCA | 1 | S241 | S241C | BRCA | 1 | F109 | Y107D | BRCA | 1 | R248 | N247I | BRCA | 1 | R280 | R280T | BRCA | 1 | K139 | C141R | BRCA | 1 | T230 | I232S | BRCA | 1 | R273 | R273L | BRCA | 1 | H168 | H168P | BRCA | 1 | G279,C277 | P278A | BRCA | 1 | R283 | R282G | BRCA | 1 | A276 | A276P | BRCA | 1 | V157 | V157F | BRCA | 1 | V157 | V157G | BRCA | 1 | L257 | I255F | BRCA | 1 | R273,C275 | V274D | COAD | 1 | C238 | C238Y | COAD | 1 | C176 | C176F | COAD | 1 | M243 | G245D | COAD | 1 | T123 | C124G | COAD | 1 | K139 | C141S | COAD | 1 | K139 | C141Y | COAD | 1 | L145 | V143A | COAD | 1 | G279,C277 | P278R | COAD | 1 | T230 | I232S | COAD | 1 | G279 | G279E | COAD | 1 | T155,P153 | G154C | COAD | 1 | P152 | P152R | COAD | 1 | G279,C277 | P278A | COAD | 1 | K139 | A138V | COAD | 1 | P151 | P151H | COAD | 1 | R273,C275 | V274L | COAD | 1 | R248 | R248Q | COAD | 1 | K139 | C141R | COAD | 1 | C238 | M237I | COAD | 1 | S241,N239 | S240R | COAD | 1 | T150 | T150A | COAD | 1 | V157 | R158C | COAD | 1 | G279 | G279W | COAD | 1 | L257 | D259V | COAD | 1 | T155 | T155I | COAD | 1 | R248 | P250L | COAD | 1 | N239 | N239D | COAD | 1 | T230 | T231I | COAD | 1 | R273,C275 | V274A | COAD | 1 | R248 | M246T | COAD | 1 | T123 | T125A | COAD | 1 | Q165,Q167 | S166L | COAD | 1 | C238 | M237L | COAD | 1 | A119 | A119V | COAD | 1 | E221 | E221G | COAD | 1 | L257 | E258G | COAD | 1 | P223 | E224D | COAD | 1 | F109 | G108C | COAD | 1 | L257 | I255T | COAD | 1 | C238 | C238R | COAD | 1 | D228 | D228G | COAD | 1 | R273 | V272G | COAD | 1 | E221 | E221V | COAD | 1 | Q165,Q167 | S166P | COAD | 1 | L257 | D259Y | COAD | 1 | V122 | V122M | COAD | 1 | F109 | F109S | COAD | 1 | C242 | C242R | COAD | 1 | T118 | S116P | COAD | 1 | R248 | M246V | COAD | 1 | C176 | H178Q | GBM | 1 | A276 | A276V | GBM | 1 | R280 | D281A | GBM | 1 | R280 | D281H | GBM | 1 | R273 | E271K | GBM | 1 | L257 | I255S | GBM | 1 | Q165 | K164E | GBM | 1 | V157 | V157G | GBM | 1 | R273 | R273C | GBM | 1 | C275 | C275Y | GBM | 1 | C242 | C242Y | GBM | 1 | R248 | R249M | GBM | 1 | R248 | M246R | GBM | 1 | R248 | R249T | GBM | 1 | P152 | P152S | GBM | 1 | M243 | G244S | GBM | 1 | T155,V157 | R156C | GBM | 1 | K139 | A138T | GBM | 1 | R248 | R248L | GBM | 1 | L257 | E258D | HNSC | 1 | T155,V157 | R156P | HNSC | 1 | C242 | C242Y | HNSC | 1 | V157 | R158L | HNSC | 1 | R273 | R273S | HNSC | 1 | C238 | M237I | HNSC | 1 | H168 | H168L | HNSC | 1 | P223 | E224D | HNSC | 1 | R248 | R249M | HNSC | 1 | M243 | G245D | HNSC | 1 | R283 | R283P | HNSC | 1 | R280 | R280T | HNSC | 1 | P151 | P151S | HNSC | 1 | V157 | R158H | HNSC | 1 | R280 | R280G | HNSC | 1 | R273 | V272M | HNSC | 1 | V157 | A159V | HNSC | 1 | C176 | C176S | HNSC | 1 | C238 | M237V | HNSC | 1 | C238 | C238S | HNSC | 1 | R280 | R280S | HNSC | 1 | C176 | C176Y | HNSC | 1 | P151 | P151T | HNSC | 1 | C242 | C242S | HNSC | 1 | Q165 | Y163C | HNSC | 1 | P151 | P151H | HNSC | 1 | R273 | E271V | HNSC | 1 | L257 | E258A | HNSC | 1 | K139 | L137Q | HNSC | 1 | C238 | Y236D | HNSC | 1 | V157 | A159P | KICH | 1 | C275 | C275Y | KICH | 1 | C176 | H178Q | KICH | 1 | N239 | N239D | KICH | 1 | K139 | C141W | KICH | 1 | R248 | M246I | KICH | 1 | R273 | R273H | KIRC | 1 | R248 | R248Q | KIRC | 1 | R280 | D281Y | KIRC | 1 | N239 | N239S | KIRC | 1 | S241 | S241C | KIRC | 1 | R273 | V272M | KIRC | 1 | C176 | C176Y | LAML | 1 | R280 | R280G | LAML | 1 | K139 | C141W | LAML | 1 | R248 | R248Q | LAML | 1 | R273 | R273C | LAML | 1 | R273 | V272G | LUAD | 1 | C176 | C176Y | LUAD | 1 | R280 | D281V | LUAD | 1 | R280 | R280I | LUAD | 1 | T155 | T155I | LUAD | 1 | R280 | D281E | LUAD | 1 | C238 | C238Y | LUAD | 1 | P151 | P151S | LUAD | 1 | F109 | L111M | LUAD | 1 | L257 | I255F | LUAD | 1 | L257 | D259Y | LUAD | 1 | T155,V157 | R156P | LUAD | 1 | C176 | P177R | LUAD | 1 | E221 | Y220C | LUAD | 1 | R248 | R249S | LUAD | 1 | R273,C275 | V274F | LUAD | 1 | S241 | S241F | LUAD | 1 | R283 | R283P | LUAD | 1 | R280 | R280T | LUAD | 1 | F109 | F109C | LUAD | 1 | M243 | G245C | LUAD | 1 | H168 | H168L | LUAD | 1 | M243 | G245V | LUAD | 1 | T230 | I232F | LUAD | 1 | R283 | E285K | LUAD | 1 | R273 | V272M | LUAD | 1 | R280 | D281Y | LUAD | 1 | L145 | Q144L | LUAD | 1 | V157 | R158P | LUAD | 1 | L145 | Q144H | LUAD | 1 | P223 | E224D | LUAD | 1 | R280 | R280G | LUAD | 1 | N239 | N239S | LUAD | 1 | T155,P153 | G154V | LUAD | 1 | R248 | R248P | LUAD | 1 | C176 | C176F | LUAD | 1 | G279,C277 | P278H | LUAD | 1 | Q165 | Y163C | LUAD | 1 | R248 | P250L | LUAD | 1 | Q165 | Y163H | LUAD | 1 | Q165 | Y163D | LUAD | 1 | R283 | R282Q | LUSC | 1 | L257 | D259V | LUSC | 1 | L257 | D259Y | LUSC | 1 | T155 | T155P | LUSC | 1 | C176 | C176Y | LUSC | 1 | P223 | E224D | LUSC | 1 | R248 | P250L | LUSC | 1 | M243 | G245C | LUSC | 1 | R273 | R273C | LUSC | 1 | C176 | C176F | LUSC | 1 | C238 | Y236C | LUSC | 1 | R248 | R248W | LUSC | 1 | R280 | R280I | LUSC | 1 | F109 | R110L | LUSC | 1 | P151 | P151R | LUSC | 1 | C238 | M237I | LUSC | 1 | Q165 | K164E | LUSC | 1 | G279,C277 | P278S | LUSC | 1 | R248 | R249W | LUSC | 1 | R248 | R248Q | LUSC | 1 | R283 | T284P | LUSC | 1 | F109 | L111Q | LUSC | 1 | R280 | D281Y | LUSC | 1 | M243 | G245S | LUSC | 1 | R283 | R282W | LUSC | 1 | R273 | R273H | LUSC | 1 | K139 | C141W | LUSC | 1 | G279,C277 | P278L | LUSC | 1 | V157 | R158H | LUSC | 1 | R248 | R248P | LUSC | 1 | T155,P153 | G154V | LUSC | 1 | M243 | G244D | LUSC | 1 | M243 | G244C | LUSC | 1 | R248 | R249S | LUSC | 1 | R273 | R273P | LUSC | 1 | F109 | L111R | LUSC | 1 | T123 | T125P | LUSC | 1 | R248 | R249G | LUSC | 1 | G279,C277 | P278A | LUSC | 1 | R248 | R248L | LUSC | 1 | V157 | R158G | LUSC | 1 | G279,C277 | P278R | LUSC | 1 | R283 | R283P | LUSC | 1 | R283 | R282P | OV | 1 | C176 | C176G | OV | 1 | L257 | L257Q | OV | 1 | Q165 | Y163H | OV | 1 | C176 | H178N | OV | 1 | A276 | A276P | OV | 1 | G279 | G279E | OV | 1 | R273 | R273P | OV | 1 | F109 | F109C | OV | 1 | C277 | C277F | OV | 1 | R280 | R280I | OV | 1 | L145 | L145Q | OV | 1 | Q165 | K164E | OV | 1 | R280 | D281G | OV | 1 | P151 | P151S | OV | 1 | T155,V157 | R156P | OV | 1 | R273,C275 | V274D | OV | 1 | N239 | N239S | OV | 1 | R248 | R249G | OV | 1 | C176 | C176F | OV | 1 | S241 | S241Y | OV | 1 | G279,C277 | P278H | OV | 1 | C176 | C176S | OV | 1 | R280 | D281V | OV | 1 | T230 | I232N | OV | 1 | R273,C275 | V274G | OV | 1 | L145 | L145R | OV | 1 | M243 | G245C | OV | 1 | T155 | T155A | OV | 1 | R248 | R248G | OV | 1 | C238 | C238G | OV | 1 | M243 | G244D | OV | 1 | R280 | D281E | OV | 1 | P151 | P151R | OV | 1 | C238 | C238F | OV | 1 | T123 | T125M | OV | 1 | K139 | A138V | OV | 1 | S241 | S241A | OV | 1 | M243 | G245R | OV | 1 | R280 | D281Y | OV | 1 | C275 | C275S | OV | 1 | C238 | C238Y | OV | 1 | Q165 | Y163N | OV | 1 | C176 | R175H | PRAD | 1 | R283 | R282W | PRAD | 1 | M243 | G245S | PRAD | 1 | N239 | N239D | PRAD | 1 | R280 | R280K | SKCM | 1 | K120 | K120E | SKCM | 1 | L145 | V143E | SKCM | 1 | G279,C277 | P278L | SKCM | 1 | C176 | P177L | SKCM | 1 | R248 | R248W | SKCM | 1 | L145 | V143G | SKCM | 1 | E221 | Y220C | SKCM | 1 | P152 | P152L | SKCM | 1 | M243 | G245R | STAD | 1 | K139 | K139N | STAD | 1 | H168 | H168R | STAD | 1 | F109 | Y107D | STAD | 1 | C238 | C238F | STAD | 1 | C176 | C176Y | STAD | 1 | R283 | E285V | STAD | 1 | R273 | V272M | STAD | 1 | P151 | P151T | STAD | 1 | F109 | L111Q | STAD | 1 | C242 | C242Y | STAD | 1 | R248 | R249S | STAD | 1 | R248 | P250L | STAD | 1 | F109 | G108S | STAD | 1 | C176 | R175C | STAD | 1 | H168 | M169T | STAD | 1 | R273 | E271K | STAD | 1 | C176 | R175G | STAD | 1 | C238 | M237I | STAD | 1 | T123 | T125M | STAD | 1 | M243 | G244C | STAD | 1 | T230 | I232T | UCEC | 1 | R273 | R273S | UCEC | 1 | P151 | P151S | UCEC | 1 | R248 | R249W | UCEC | 1 | C176 | H178D | UCEC | 1 | R273 | V272M | UCEC | 1 | C238 | C238F | UCEC | 1 | R283 | R282W | UCEC | 1 | C238 | C238Y | UCEC | 1 | N239 | N239D | UCEC | 1 | R280 | D281Y | UCEC | 1 | K139 | C141Y | UCEC | 1 | N239 | N239S | UCEC | 1 | S241 | S241P | UCEC | 1 | R248 | R249S | UCEC | 1 | C238 | M237K | UCEC | 1 | S241 | S241C | UCEC | 1 | G279,C277 | P278T | UCEC | 1 | S241 | S241F | UCEC | 1 |
| cf) Cancer type abbreviation. BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, COAD: Colon adenocarcinoma, GBM: Glioblastoma multiforme, LGG: Brain lower grade glioma, HNSC: Head and neck squamous cell carcinoma, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute myeloid leukemia, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PRAD: Prostate adenocarcinoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, THCA: Thyroid carcinoma, UCEC: Uterine corpus endometrial carcinoma. |
 Clinical information for TP53 from My Cancer Genome. Clinical information for TP53 from My Cancer Genome. |
| Tumor protein p53 (TP53) is a gene that codes for a tumor suppressor protein. The protein regulates expression of genesinvolved in cell cycle arrest, apoptosis, senescence, DNA repair, and changes in metabolism (Gene 2014). In cancer, TP53’s normal roles are not fulfilled, leading to cell survival, DNA damage, and cell proliferation. TP53 is the most frequently mutated gene in cancer; it is mutated in about half of all cancers (Genetics Home Reference 2014). TP53 is most frequently mutated in ovarian, colon, and esophageal cancers, although it is significantly mutated in many other cancer types (COSMIC). Strickland, S.A., Kim, A.S. 2015. TP53. My Cancer Genomehttps://www.mycancergenome.org/content/gene/tp53/ (Updated December 2015) |
| Top |
| Protein structure related information for TP53 |
 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Relative protein structure stability change (ΔΔE) using Mupro 1.1 Mupro score denotes assessment of the effect of mutations on thermodynamic stability. (ΔΔE<0: mutation decreases stability, ΔΔE>0: mutation increases stability) |
 : nsSNV at non-LBS : nsSNV at non-LBS : nsSNV at LBS : nsSNV at LBS |
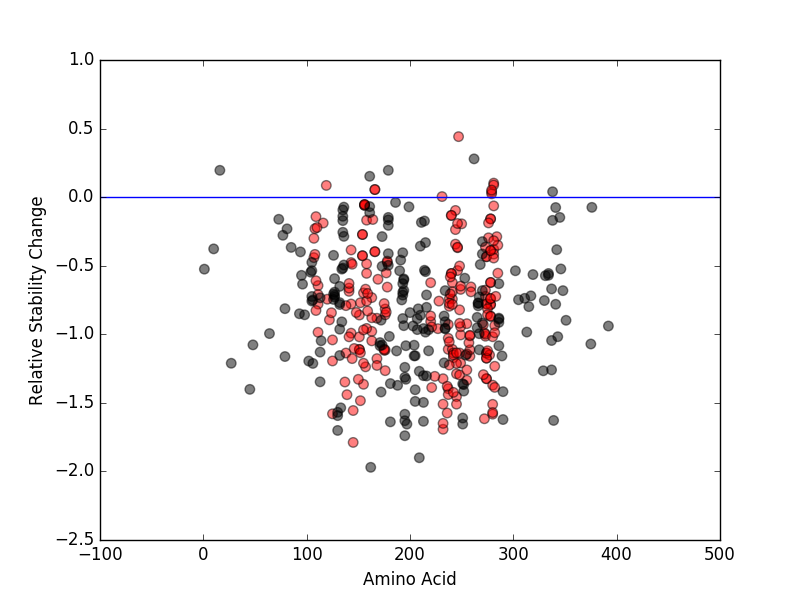 |
 nsSNVs sorted by the relative stability change of protein structure by each mutation nsSNVs sorted by the relative stability change of protein structure by each mutation Blue: mutations of positive stability change. and red : the most recurrent mutation for this gene. |
| LBS | AAchange of nsSNV | Relative stability change | R248 | N247I | 0.44123146 | R280 | D281V | 0.10228051 | R280 | D281E | 0.087914822 | A119 | A119V | 0.085624008 | Q167 | S166L | 0.055371255 | Q165 | S166L | 0.055371255 | G279 | G279R | 0.051401998 | G279 | G279W | 0.038815426 | G279 | G279E | 0.02424754 | T230 | T231I | 0.004029671 | L145 | L145Q | -1.7884843 | T230 | I232N | -1.6927216 | T230 | I232S | -1.6496924 | R273 | V272G | -1.6159679 | R280 | R280G | -1.5823802 | T123 | T125A | -1.5799886 | C238 | Y236D | -1.5741252 | R280 | R280S | -1.5705052 | L145 | L145R | -1.5558493 | R280 | R280K | -1.5106393 | T230 | I232T | -1.5095945 | M243 | G245S | -1.5093679 | P152 | P152S | -1.4842484 | M243 | G245D | -1.454748 | K139 | K139N | -1.4403836 | C238 | M237T | -1.4395932 | C242 | C242S | -1.4236268 | C238 | M237K | -1.3967598 | E221 | E221G | -1.3882277 | R283 | R282G | -1.3872027 | C238 | Y236C | -1.379064 | R280 | R280T | -1.3709839 | T155 | T155N | -1.3646021 | K139 | L137Q | -1.3479875 | L257 | I255N | -1.335189 | T150 | T150A | -1.3282886 | T230 | I232F | -1.3239524 | R273 | V274G | -1.3238858 | C275 | V274G | -1.3238858 | P223 | E224D | -1.3067282 | R248 | R248G | -1.2964135 | R273 | E271K | -1.2943414 | M243 | G245C | -1.2881699 | C176 | C176G | -1.2652152 | L257 | I255S | -1.2601318 | V157 | V157G | -1.2372881 | R283 | R282Q | -1.2332047 | H168 | H168P | -1.2270711 | C238 | C238G | -1.2241451 | M243 | G245V | -1.2130118 | T155 | T155A | -1.2111341 | R248 | R249G | -1.2094495 | T123 | T125P | -1.1945216 | H168 | H168R | -1.1775171 | L145 | Q144H | -1.177424 | C275 | V274D | -1.1739382 | R273 | V274D | -1.1739382 | C242 | C242Y | -1.1711171 | L257 | E258G | -1.1599803 | C242 | C242F | -1.151124 | C277 | C277F | -1.1504446 | L257 | I255T | -1.1432196 | R273 | V274A | -1.1370788 | C275 | V274A | -1.1370788 | K139 | A138T | -1.136694 | C242 | C242R | -1.1365082 | R248 | R248Q | -1.136464 | M243 | G245R | -1.1354599 | P152 | P152R | -1.1345378 | C275 | C275S | -1.1202084 | C176 | R175G | -1.1166628 | C176 | C176S | -1.1152157 | L257 | E258A | -1.1129563 | P151 | P151H | -1.1117864 | P151 | P151S | -1.1101802 | C238 | M237V | -1.1095958 | S241 | S241A | -1.1094179 | C176 | R175H | -1.1087266 | W146 | W146S | -1.1035714 | L257 | L257Q | -1.0616103 | C238 | C238S | -1.0545455 | Q165 | Y163N | -1.0478075 | T123 | T125M | -1.0410024 | C238 | M237I | -1.0291636 | P151 | P151T | -1.0187024 | K139 | C141S | -1.0185365 | R280 | R280I | -1.0178892 | R273 | R273S | -1.014134 | L257 | E258Q | -1.0114936 | L257 | E258D | -1.0073882 | R248 | R249S | -1.0047248 | R273 | R273H | -0.99942546 | L145 | V143G | -0.99170842 | R273 | V272M | -0.9901801 | R283 | R282P | -0.98854452 | F109 | L111Q | -0.98536048 | Q165 | Y163H | -0.97893475 | T155 | T155P | -0.96836792 | D228 | S227P | -0.95912427 | V157 | R158G | -0.95726327 | E221 | E221V | -0.9463474 | R248 | R249T | -0.94470639 | R283 | R282W | -0.93664617 | R273 | E271V | -0.92991765 | C238 | M237L | -0.92890116 | L257 | I255F | -0.92265071 | R248 | R248P | -0.91996727 | E221 | Y220H | -0.90727385 | V122 | V122M | -0.89472201 | H168 | H168L | -0.88311906 | Q165 | Y163D | -0.87952435 | G279 | P278A | -0.86850193 | C277 | P278A | -0.86850193 | E221 | Y220S | -0.86650363 | P151 | P151R | -0.85987578 | C176 | C176Y | -0.85829507 | C275 | C275Y | -0.85686094 | T123 | C124G | -0.8430408 | D148 | D148N | -0.84109856 | C176 | P177R | -0.83831643 | V157 | A159P | -0.83021755 | F109 | F109S | -0.82666078 | R248 | R248W | -0.82181901 | C176 | C176F | -0.81576335 | C275 | C275F | -0.81420142 | C238 | C238Y | -0.79586672 | K139 | A138V | -0.79101398 | C277 | P278S | -0.78324005 | G279 | P278S | -0.78324005 | S241 | S241C | -0.78241232 | F109 | L111P | -0.78003926 | L145 | V143A | -0.77927069 | R273 | V274F | -0.77832623 | C275 | V274F | -0.77832623 | C176 | R175C | -0.77610417 | C238 | C238F | -0.77315204 | P152 | P152L | -0.77011045 | D228 | D228G | -0.75814898 | V157 | R158H | -0.75779946 | K120 | K120E | -0.74438529 | S241 | S241P | -0.74294193 | C238 | C238R | -0.74135505 | G279 | P278H | -0.73940287 | C277 | P278H | -0.73940287 | Q165 | Y163C | -0.73263737 | R283 | R283P | -0.72348885 | R273 | R273P | -0.72309185 | F109 | L111R | -0.71044699 | N239 | N239D | -0.70606751 | V157 | A159V | -0.70220185 | T155 | T155I | -0.69869425 | L257 | D259Y | -0.69045917 | S241 | S241Y | -0.6896448 | K139 | C141Y | -0.68687583 | R273 | R273C | -0.67808717 | R248 | R249W | -0.6725509 | V157 | V157F | -0.67113294 | K139 | C141R | -0.66313343 | C176 | H178N | -0.65581918 | L257 | D259V | -0.65454798 | R248 | R249M | -0.64848408 | F109 | L111M | -0.64495342 | K139 | C141W | -0.62940634 | E221 | Y220C | -0.62388851 | S241 | S241F | -0.62343298 | G279 | P278T | -0.62232062 | C277 | P278T | -0.62232062 | F109 | F109C | -0.60954138 | H168 | M169T | -0.59891235 | N239 | N239S | -0.58212119 | N239 | S240G | -0.55668104 | S241 | S240G | -0.55668104 | V157 | R158P | -0.55570515 | R283 | E285K | -0.55445162 | C176 | P177L | -0.55260316 | R248 | M246T | -0.53525744 | R248 | R248L | -0.5022807 | L145 | Q144L | -0.48926379 | V157 | R158C | -0.48647949 | L145 | V143E | -0.47721026 | C176 | H178Q | -0.47555968 | C176 | H178D | -0.46648234 | R280 | D281G | -0.44400347 | F109 | Y107D | -0.44103265 | C275 | V274L | -0.43449135 | R273 | V274L | -0.43449135 | P153 | G154C | -0.42710508 | T155 | G154C | -0.42710508 | F109 | G108S | -0.41753493 | R280 | D281A | -0.41528159 | Q167 | S166P | -0.39779246 | Q165 | S166P | -0.39779246 | C277 | P278R | -0.38951953 | G279 | P278R | -0.38951953 | L145 | V143M | -0.38499489 | R280 | D281N | -0.38091774 | R248 | M246V | -0.36884508 | R248 | M246R | -0.36502092 | R273 | R273L | -0.36417523 | R283 | E285V | -0.35028933 | M243 | G244S | -0.34284316 | R280 | D281H | -0.31768233 | F109 | Y107C | -0.30028122 | A276 | A276P | -0.29665064 | R283 | T284P | -0.28832358 | T155 | G154V | -0.2724623 | P153 | G154V | -0.2724623 | M243 | G244D | -0.23648851 | F109 | G108C | -0.22972074 | F109 | R110L | -0.22154768 | R248 | P250L | -0.19459107 | R248 | M246I | -0.19226039 | T118 | S116P | -0.1888892 | A276 | A276V | -0.18794386 | V157 | R158L | -0.16781574 | Q165 | K164E | -0.16295945 | C277 | P278L | -0.15853097 | G279 | P278L | -0.15853097 | F109 | F109L | -0.14215741 | N239 | S240R | -0.13328368 | S241 | S240R | -0.13328368 | M243 | G244C | -0.097008628 | R280 | D281Y | -0.063842998 | T155 | R156C | -0.059604549 | V157 | R156C | -0.059604549 | T155 | R156P | -0.053761091 | V157 | R156P | -0.053761091 |
| (MuPro1.1: Jianlin Cheng et al., Prediction of Protein Stability Changes for Single-Site Mutations Using Support Vector Machines, PROTEINS: Structure, Function, and Bioinformatics. 2006, 62:1125-1132) |
 Structure image for TP53 from PDB Structure image for TP53 from PDB |
| PDB ID | PDB title | PDB structure | 1DT7 | SOLUTION STRUCTURE OF THE C-TERMINAL NEGATIVE REGULATORY DOMAIN OF P53 IN A COMPLEX WITH CA2+-BOUND S100B(BB) | 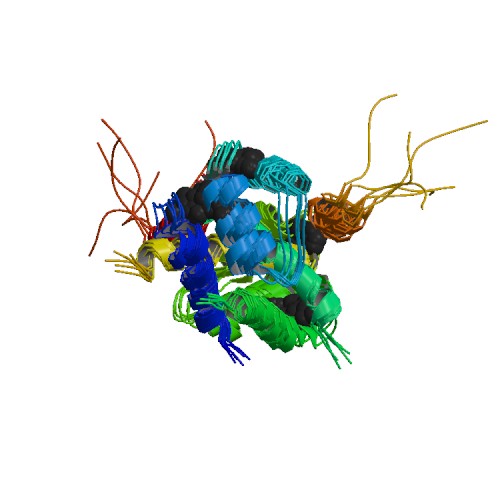 |
| Top |
| Differential gene expression and gene-gene network for TP53 |
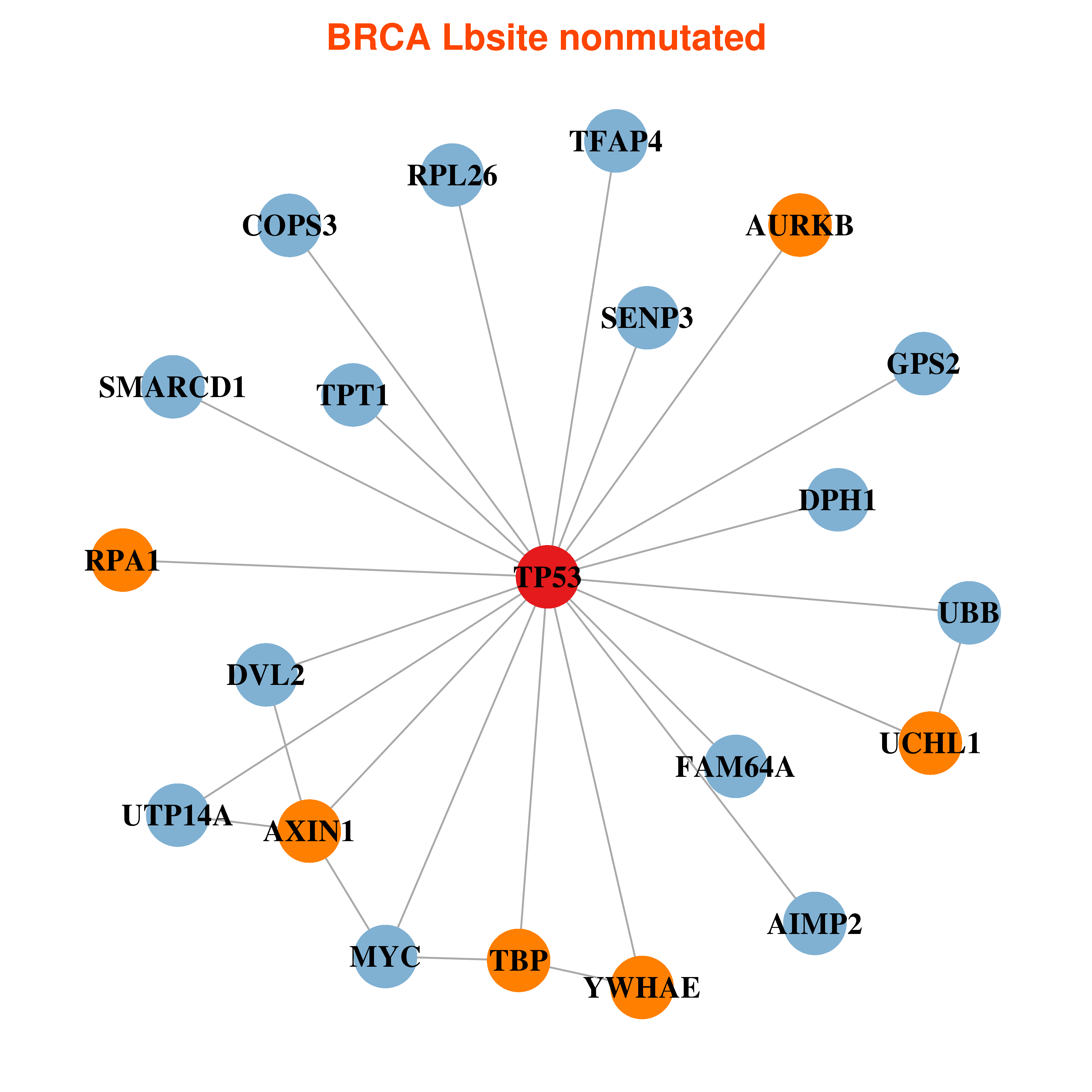
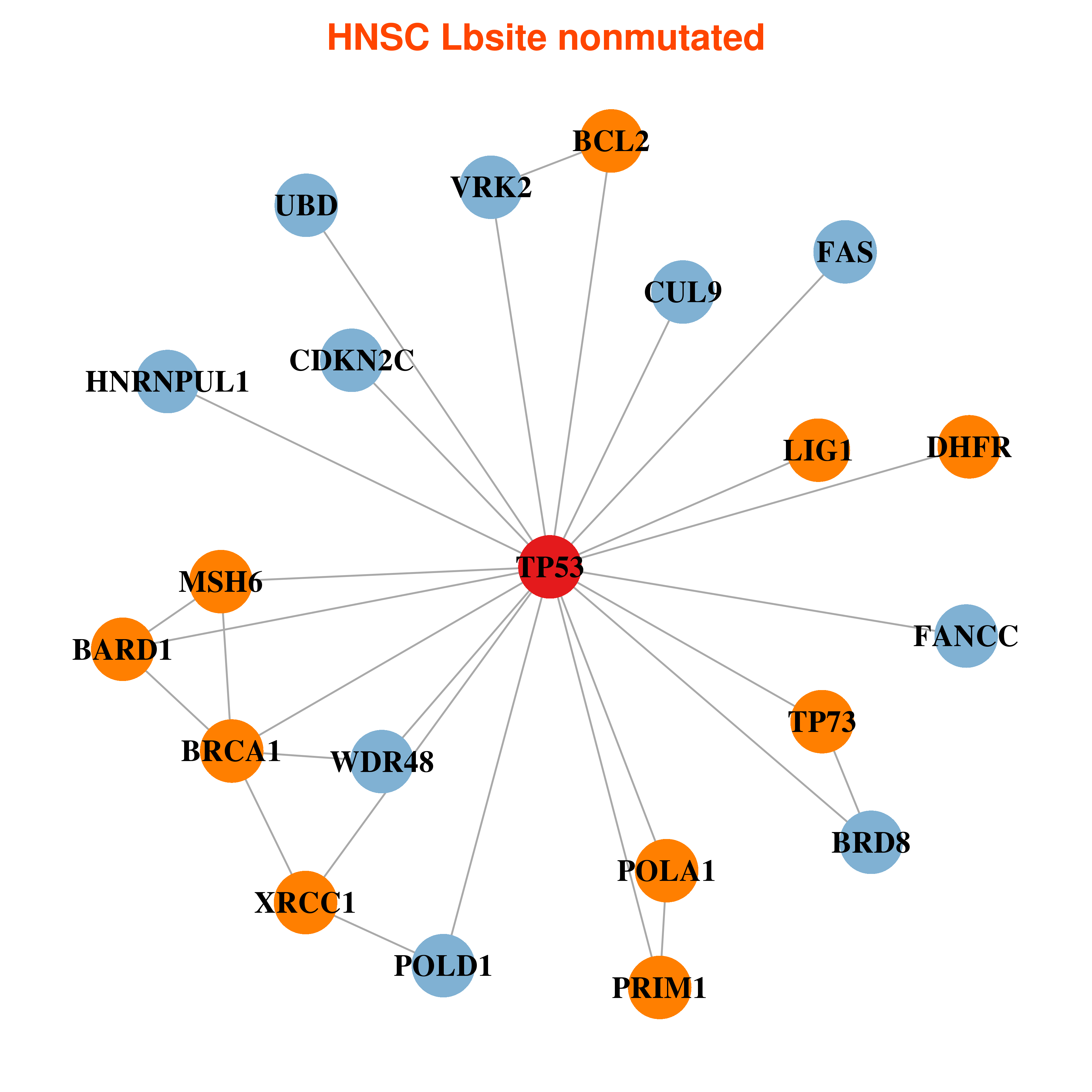
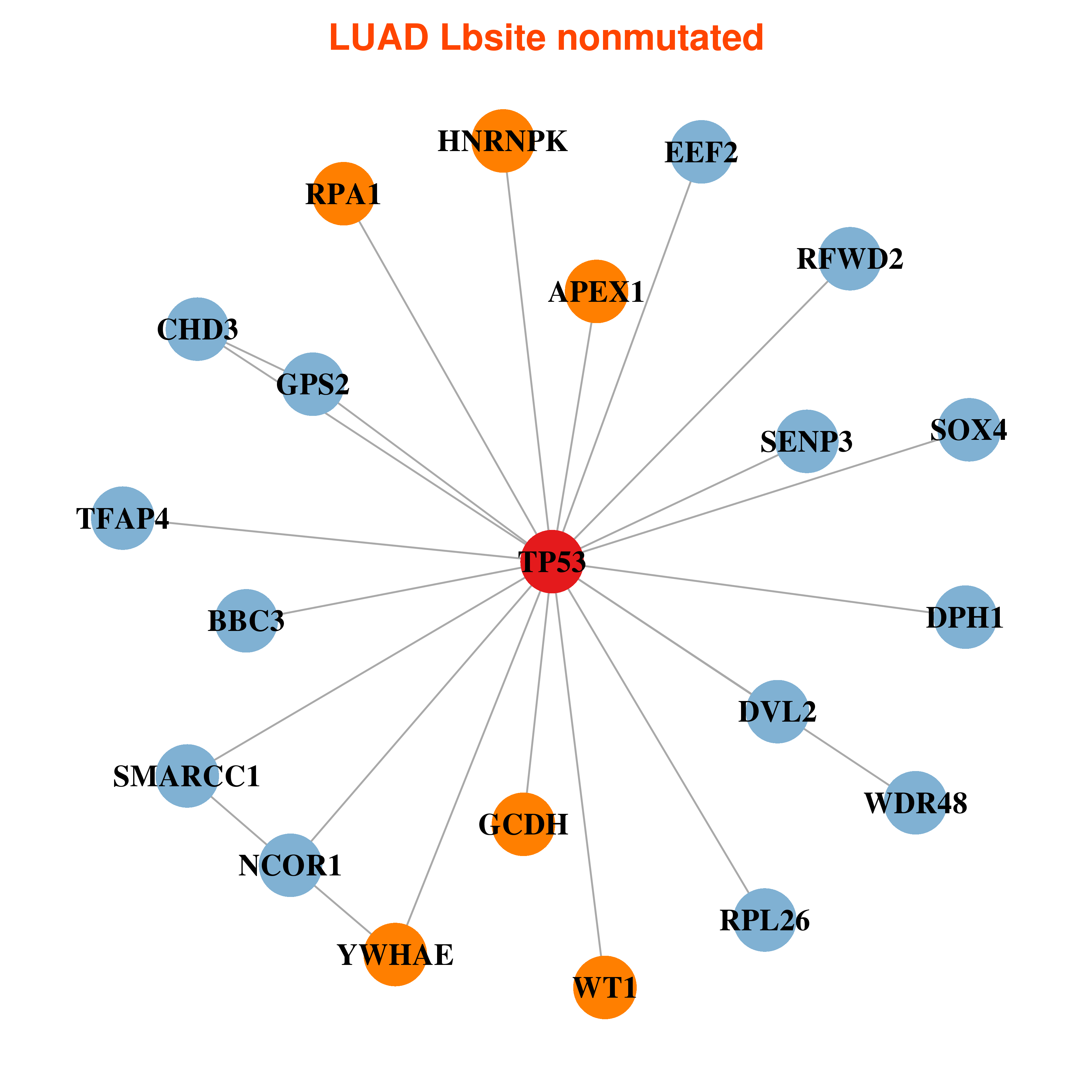
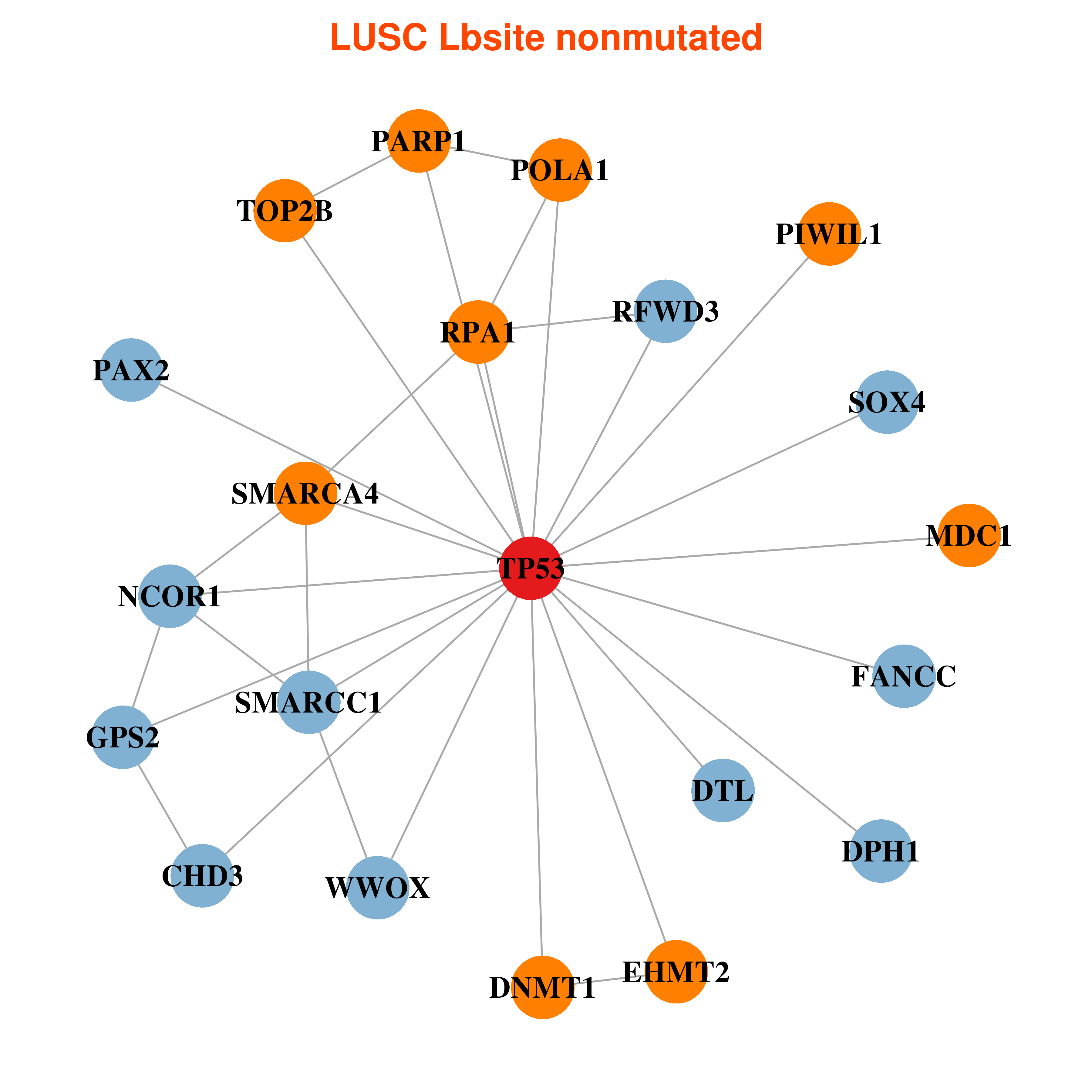
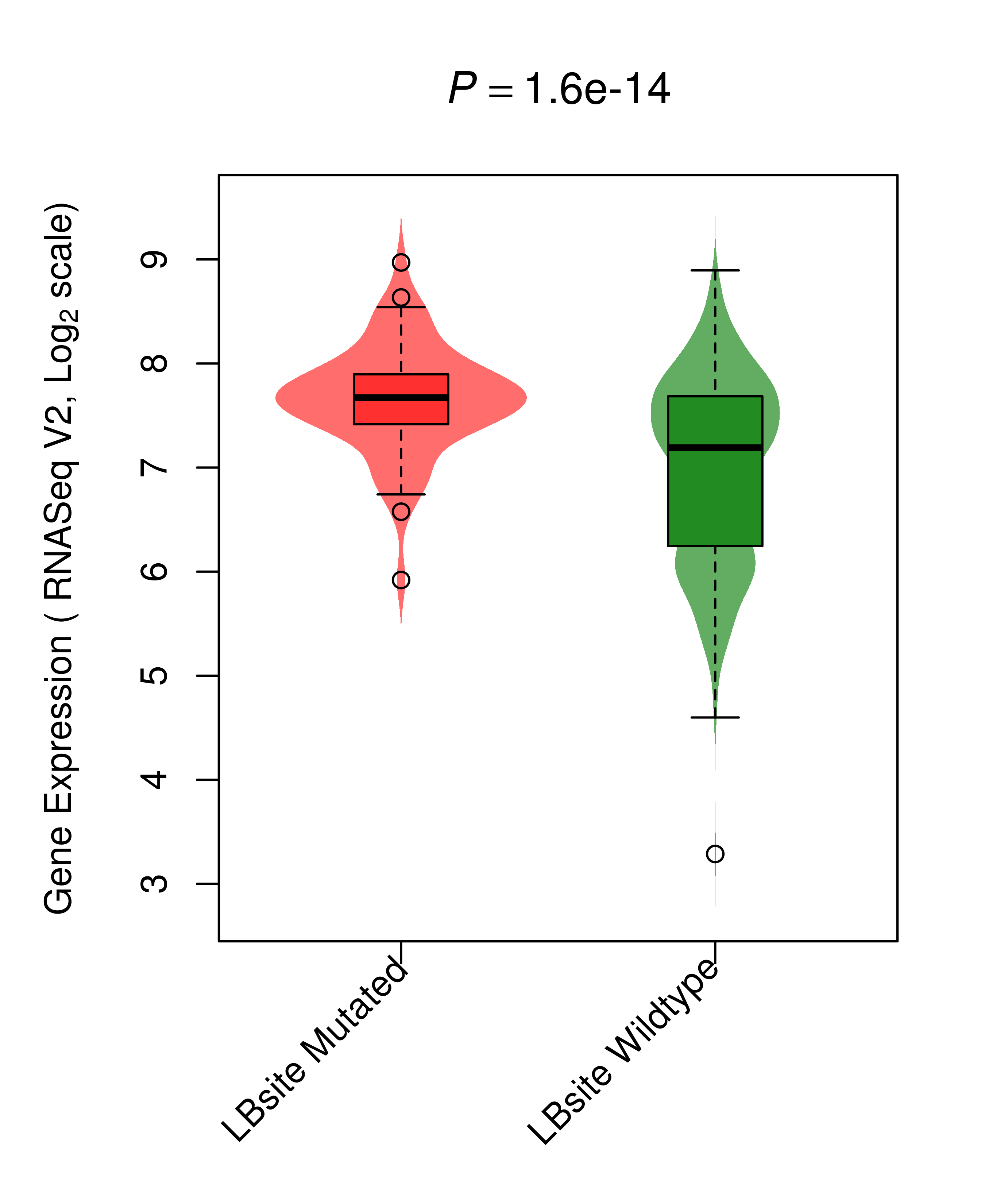
 Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types |
| TP53_BRCA_DE |
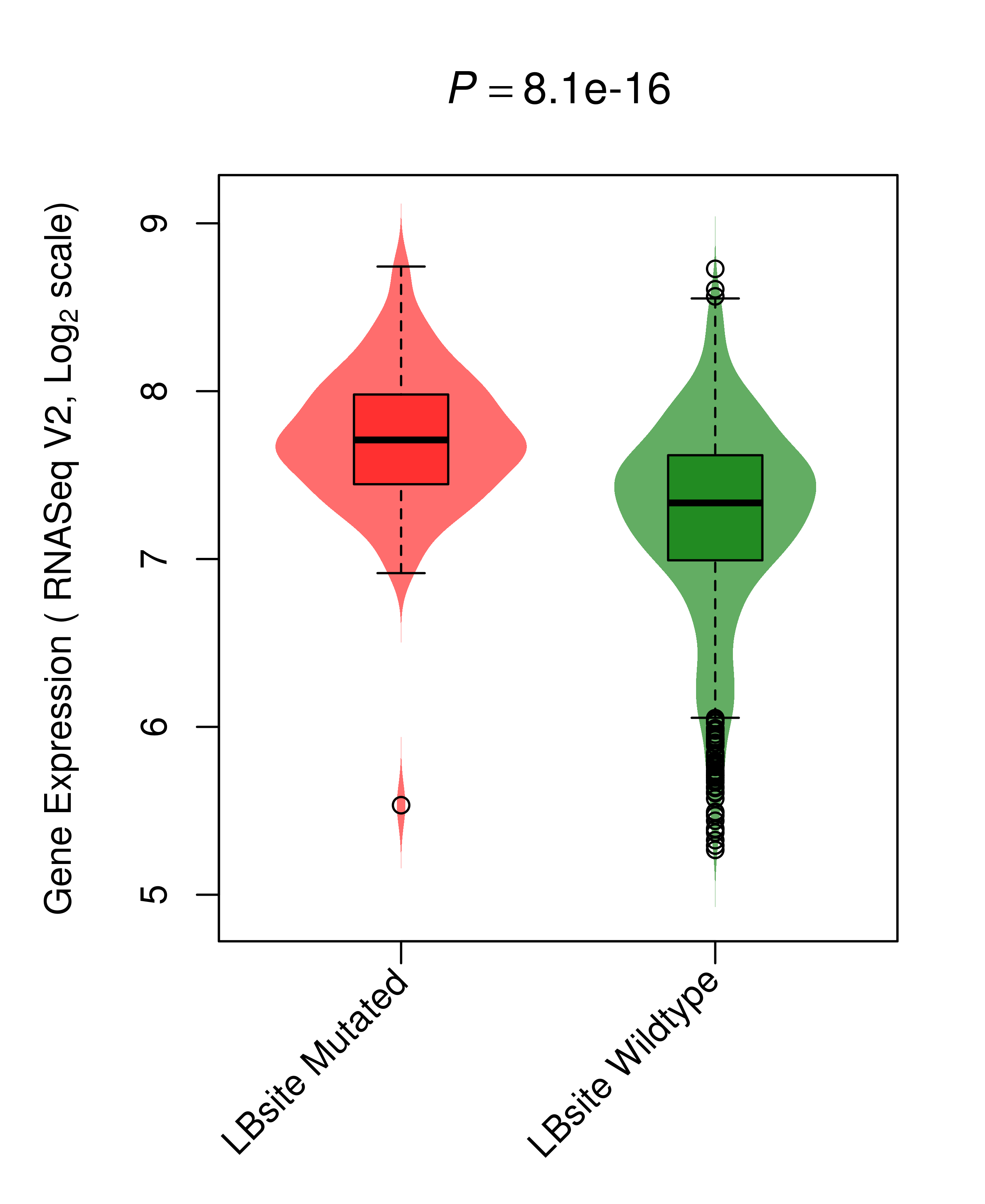 |
| TP53_COAD_DE |
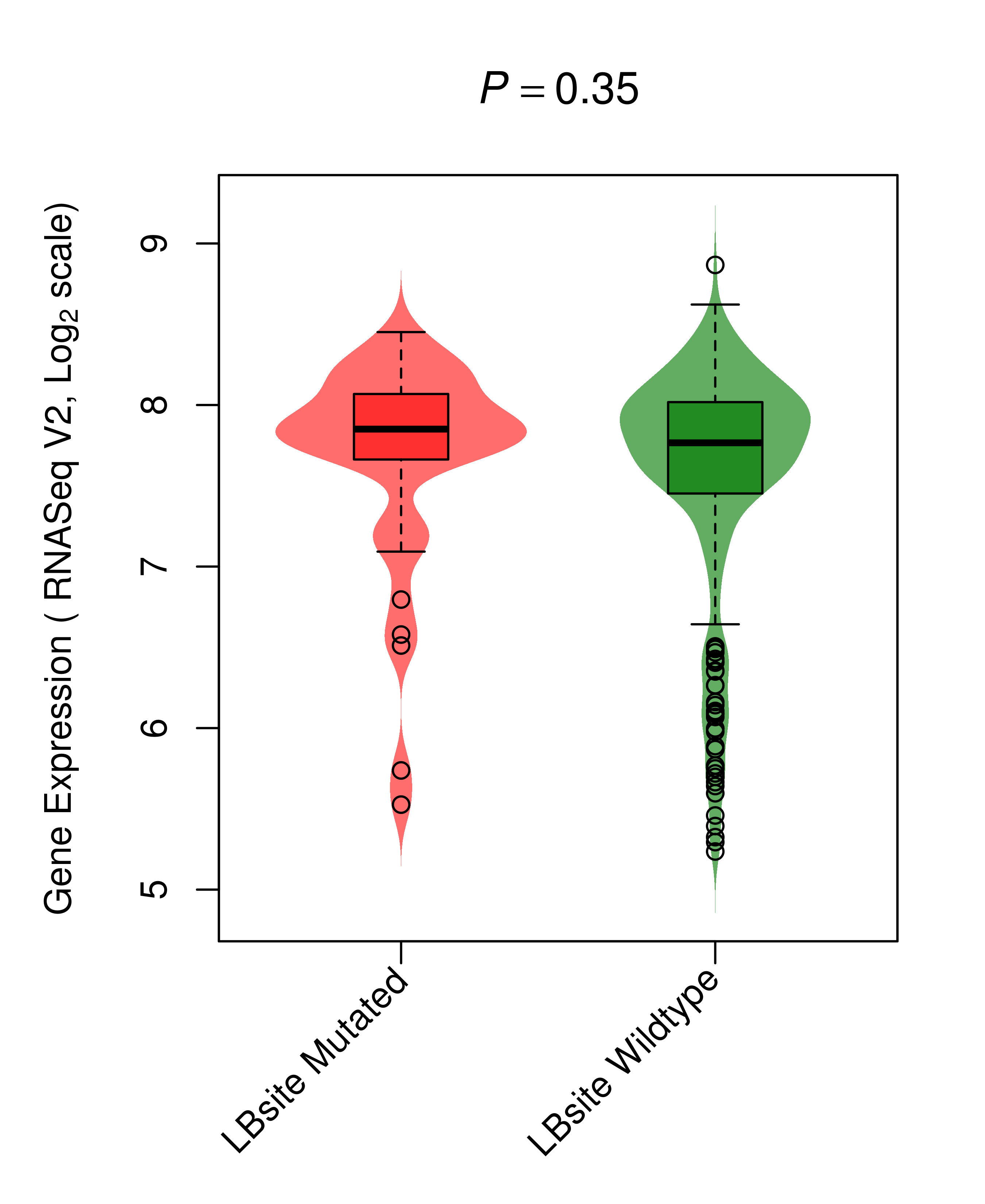 |
| TP53_HNSC_DE |
 |
| TP53_LUAD_DE |
 |
| TP53_LUSC_DE |
 |
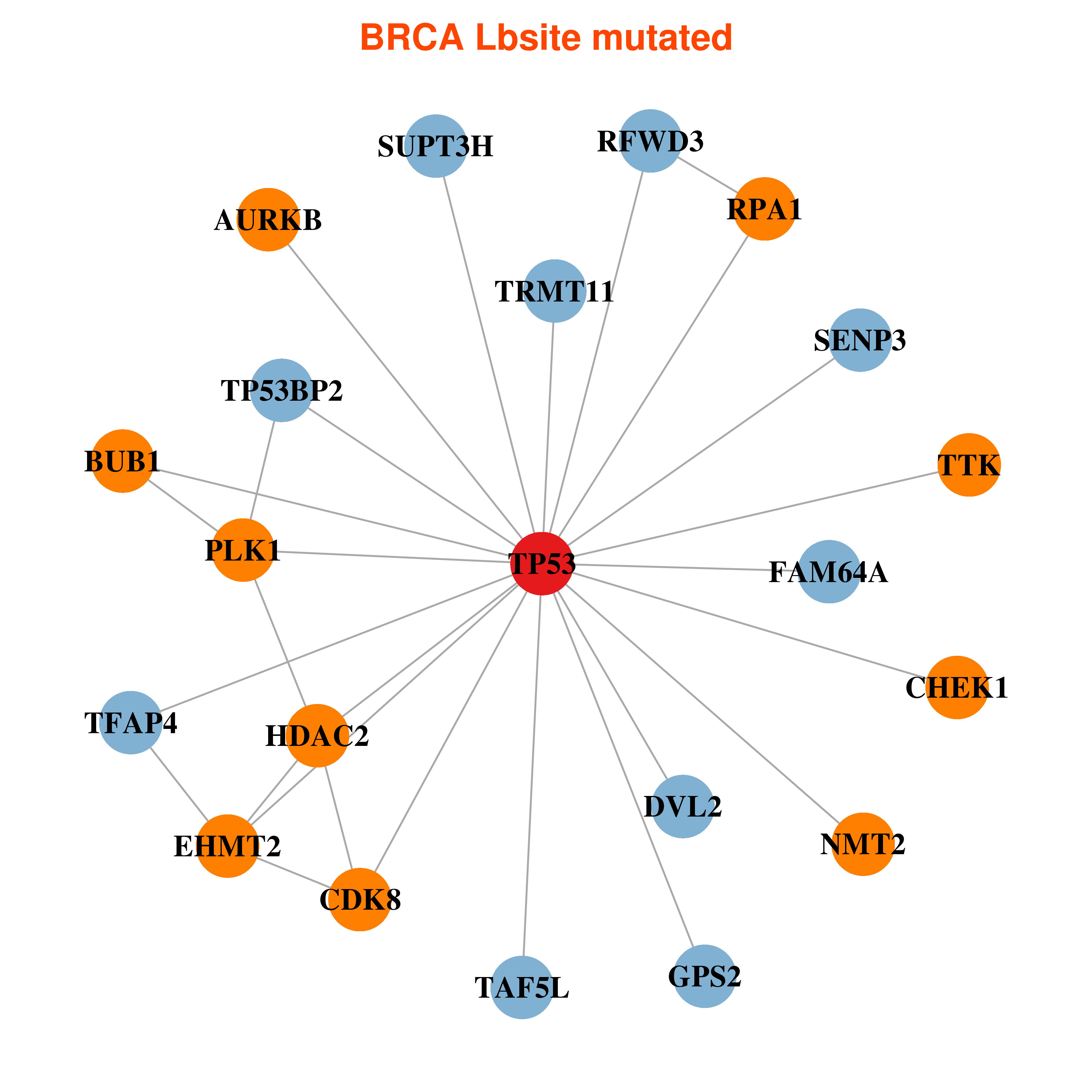
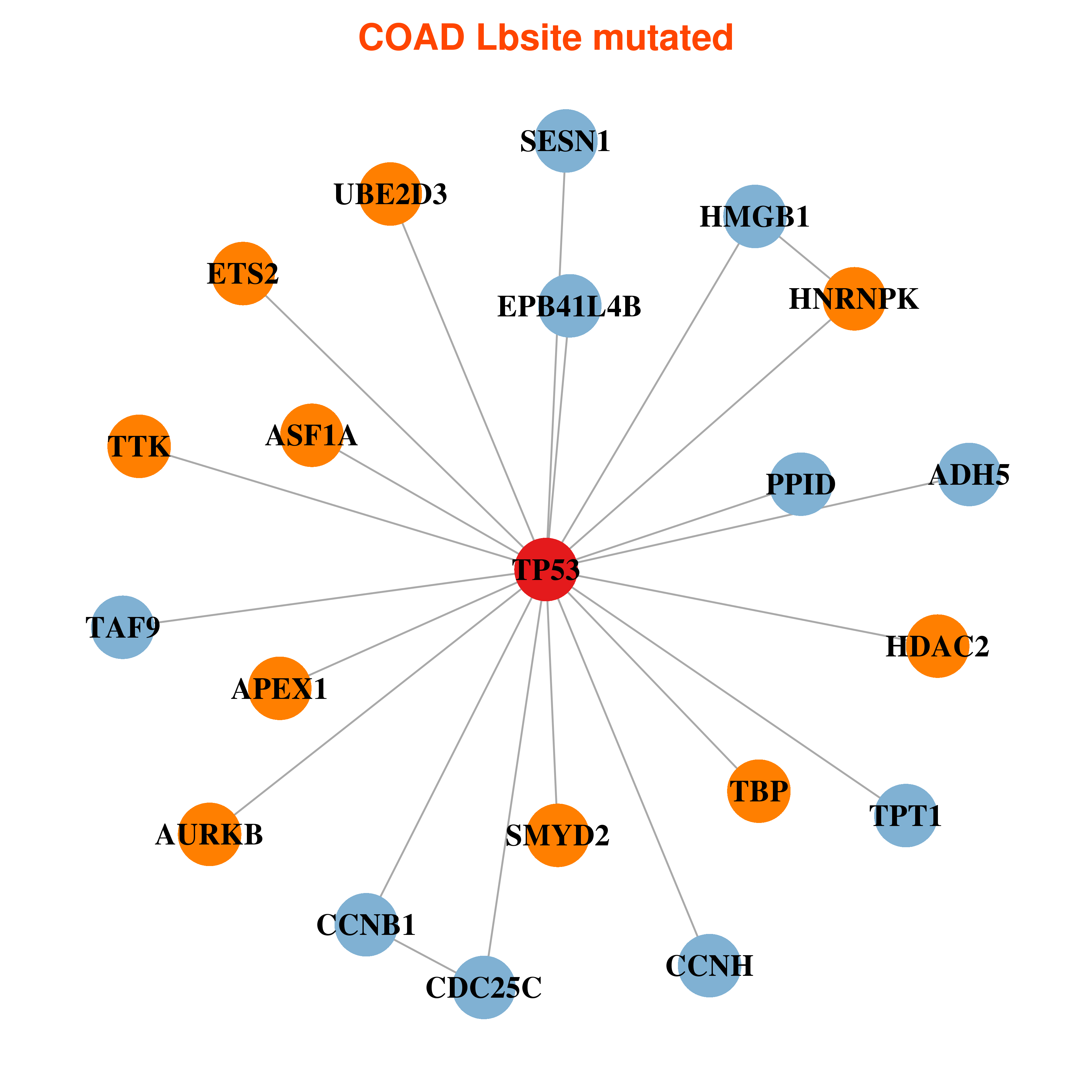
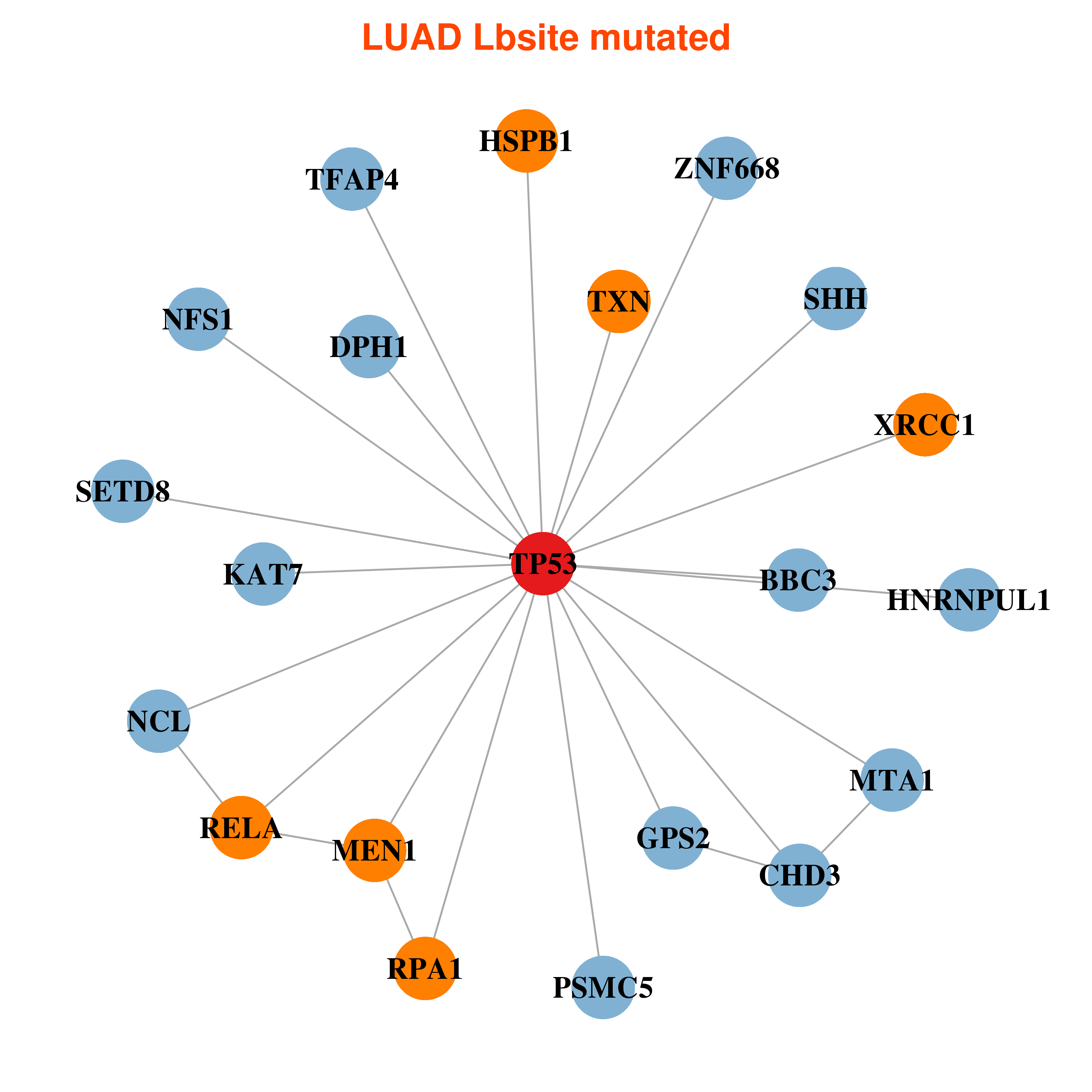
 Differential co-expressed gene network based on protein-protein interaction data (CePIN) Differential co-expressed gene network based on protein-protein interaction data (CePIN) |
| * In BRCA | |
 |
|
| * In COAD | |
 |
|
| * In HNSC | |
 |
|
| * In LUAD | |
 |
|
| * In LUSC | |
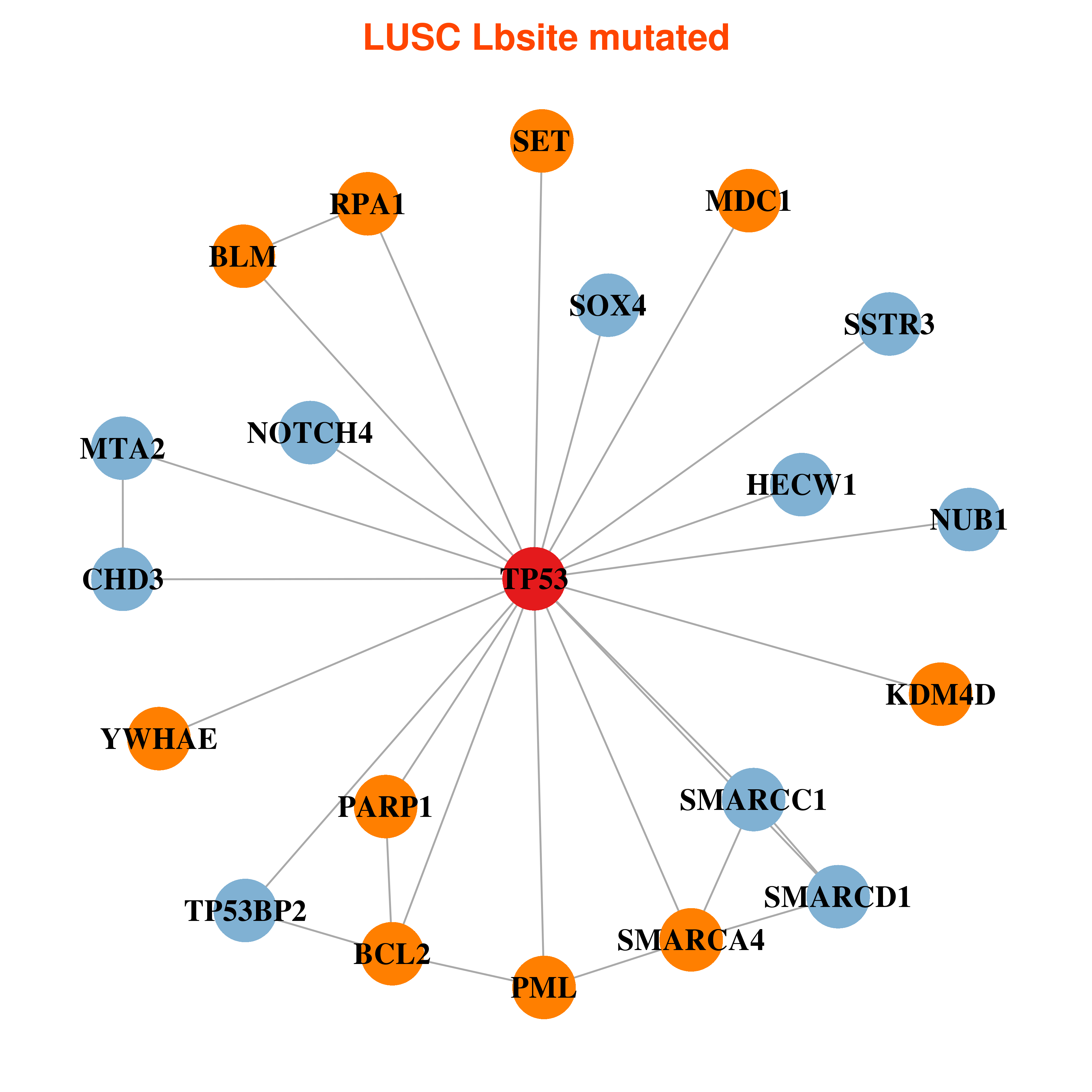 |
|
| Top |
| Top |
| Phenotype information for TP53 |
 Gene level disease information (DisGeNet) Gene level disease information (DisGeNet) |
| Disease ID | Disease name | # PubMed | Association type |
| umls:C0007097 | Carcinoma | 928 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0007137 | Carcinoma, Squamous Cell | 622 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C2239176 | Carcinoma, Hepatocellular | 497 | AlteredExpression, Biomarker, GeneticVariation, Therapeutic |
| umls:C0027627 | Neoplasm Metastasis | 391 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0001418 | Adenocarcinoma | 346 | AlteredExpression, Biomarker, GeneticVariation, Therapeutic |
| umls:C0007131 | Carcinoma, Non-Small-Cell Lung | 318 | Biomarker, GeneticVariation |
| umls:C0085390 | Li-Fraumeni Syndrome | 282 | Biomarker, GeneticVariation |
| umls:C0017638 | Glioma | 267 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0302592 | Cervix carcinoma | 260 | Biomarker, GeneticVariation |
| umls:C1458155 | Breast Neoplasms | 237 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0023434 | Leukemia, Lymphocytic, Chronic, B-Cell | 219 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0025202 | Melanoma | 196 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C1168401 | Carcinoma, squamous cell of head and neck | 170 | Biomarker, GeneticVariation |
| umls:C1261473 | Sarcoma | 169 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0001430 | Adenoma | 147 | Biomarker, GeneticVariation |
| umls:C0024121 | Lung Neoplasms | 146 | AlteredExpression, Biomarker, GeneticVariation, Therapeutic |
| umls:C0004114 | Astrocytoma | 138 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0029463 | Osteosarcoma | 135 | Biomarker, GeneticVariation, Therapeutic |
| umls:C0235974 | Pancreatic Carcinoma | 122 | Biomarker, GeneticVariation |
| umls:C0014859 | Esophageal Neoplasms | 118 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0009404 | Colorectal Neoplasms | 96 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0007138 | Carcinoma, Transitional Cell | 95 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0279626 | Esophageal Squamous Cell Carcinoma | 93 | Biomarker, GeneticVariation |
| umls:C0023903 | Liver Neoplasms | 89 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0005695 | Urinary Bladder Neoplasms | 80 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0007134 | Carcinoma, Renal Cell | 76 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C2931822 | Nasopharyngeal carcinoma | 66 | Biomarker, GeneticVariation |
| umls:C0009375 | Colonic Neoplasms | 63 | AlteredExpression, Biomarker, GeneticVariation, PostTranslationalModification |
| umls:C0037286 | Skin Neoplasms | 58 | Biomarker, GeneticVariation |
| umls:C0038356 | Stomach Neoplasms | 55 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0149925 | Small Cell Lung Carcinoma | 54 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C1961102 | Precursor Cell Lymphoblastic Leukemia-Lymphoma | 53 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0206686 | Adrenocortical Carcinoma | 49 | Biomarker, GeneticVariation |
| umls:C0040136 | Thyroid Neoplasms | 45 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0033578 | Prostatic Neoplasms | 38 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0279628 | Adenocarcinoma Of Esophagus | 36 | Biomarker, GeneticVariation |
| umls:C0030297 | Pancreatic Neoplasms | 31 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0026640 | Mouth Neoplasms | 28 | Biomarker, GeneticVariation |
| umls:C0206698 | Cholangiocarcinoma | 28 | AlteredExpression, Biomarker |
| umls:C1835398 | Sarcoma family syndrome of Li and Fraumeni | 20 | Biomarker, GeneticVariation |
| umls:C2675080 | Li-Fraumeni-Like Syndrome | 18 | Biomarker, GeneticVariation |
| umls:C0262584 | Carcinoma, Small Cell | 15 | Biomarker, PostTranslationalModification |
| umls:C0018923 | Hemangiosarcoma | 13 | Biomarker |
| umls:C0010606 | Carcinoma, Adenoid Cystic | 13 | Biomarker |
| umls:C0036341 | Schizophrenia | 12 | Biomarker, GeneticVariation |
| umls:C0024305 | Lymphoma, Non-Hodgkin | 12 | Biomarker, GeneticVariation |
| umls:C0033860 | Psoriasis | 11 | Biomarker, GeneticVariation |
| umls:C0001624 | Adrenal Gland Neoplasms | 11 | Biomarker |
| umls:C0431109 | Choroid Plexus Carcinoma | 10 | Biomarker, GeneticVariation |
| umls:C0007117 | Carcinoma, Basal Cell | 9 | AlteredExpression, Biomarker, GeneticVariation |
| umls:C0022593 | Keratosis | 9 | Biomarker, GeneticVariation |
| umls:C0022116 | Ischemia | 8 | Biomarker |
| umls:C0206681 | Adenocarcinoma, Clear Cell | 8 | Biomarker, GeneticVariation |
| umls:C0007621 | Cell Transformation, Neoplastic | 7 | Biomarker, GeneticVariation |
| umls:C0040100 | Thymoma | 6 | Biomarker |
| umls:C0858252 | Breast Adenocarcinoma | 6 | Biomarker, GeneticVariation |
| umls:C0024117 | Pulmonary Disease, Chronic Obstructive | 5 | Biomarker, GeneticVariation |
| umls:C0004698 | Balkan Nephropathy | 5 | Biomarker, GeneticVariation |
| umls:C0002736 | Amyotrophic Lateral Sclerosis | 5 | Biomarker |
| umls:C0016978 | Gallbladder Neoplasms | 4 | Biomarker, GeneticVariation |
| umls:C0027672 | Neoplastic Syndromes, Hereditary | 4 | Biomarker, GeneticVariation |
| umls:C0205770 | Papilloma, Choroid Plexus | 3 | Biomarker, GeneticVariation |
| umls:C0021361 | Infertility, Female | 3 | Biomarker, GeneticVariation |
| umls:C0030849 | Penile Neoplasms | 3 | Biomarker, GeneticVariation |
| umls:C0021364 | Infertility, Male | 3 | Biomarker, GeneticVariation |
| umls:C0311375 | Arsenic Poisoning | 3 | Biomarker, GeneticVariation |
| umls:C0007273 | Carotid Artery Diseases | 2 | Biomarker, GeneticVariation |
| umls:C0042065 | Urogenital Neoplasms | 2 | Biomarker, GeneticVariation |
| umls:C0020538 | Hypertension | 2 | Biomarker |
| umls:C0022783 | Vulvar Lichen Sclerosus | 2 | Biomarker |
| umls:C2609414 | Acute Kidney Injury | 1 | Biomarker |
| umls:C0007786 | Brain Ischemia | 1 | Biomarker |
| umls:C0007194 | Cardiomyopathy, Hypertrophic | 1 | Biomarker |
| umls:C2931713 | Chromosome 17 deletion | 1 | Biomarker |
| umls:C0011303 | Demyelinating Diseases | 1 | Biomarker |
| umls:C0013990 | Emphysema | 1 | Biomarker |
| umls:C1860789 | Leukemia, Megakaryoblastic, of Down Syndrome | 1 | Biomarker |
| umls:C0023897 | Liver Diseases, Parasitic | 1 | Biomarker |
| umls:C0023904 | Liver Neoplasms, Experimental | 1 | Biomarker |
| umls:C0042076 | Urologic Neoplasms | 1 | Biomarker |
 Mutation level pathogenic information (ClinVar annotation) Mutation level pathogenic information (ClinVar annotation) |
| Allele ID | AA change | Clinical significance | Origin | Phenotype IDs |
| 27386 | R248W | Likely pathogenic;Pathogenic | Germline;unknown | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 MedGen:C0027672 SNOMED CT:699346009 |
| 27388 | G245C | Pathogenic | Germline | GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 MedGen:C0027672 SNOMED CT:699346009 |
| 27391 | R249S | Pathogenic | Somatic | MedGen:C0302592 OMIM:603956 SNOMED CT:285432005 MedGen:C2239176 OMIM:114550 SNOMED CT:187769009 SNOMED CT:25370001 |
| 27392 | V157F | Pathogenic | Somatic | MedGen:C2239176 OMIM:114550 SNOMED CT:187769009 SNOMED CT:25370001 |
| 27393 | C242Y | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 MedGen:C2675080 |
| 27394 | G245D | Pathogenic | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 MedGen:C0027672 SNOMED CT:699346009 |
| 27395 | R248Q | Pathogenic | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 MedGen:C0027672 SNOMED CT:699346009 MedGen:C1261473 MedGen:C1836482 OMIM:609265 Orphanet:ORPHA524 |
| 27398 | S241F | Likely pathogenic;Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 MedGen:C0029463 OMIM:259500 Orphanet:ORPHA668 SNOMED CT:21708004 MedGen:C0206624 |
| 27403 | R282W | Likely benign;Pathogenic | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 MedGen:C0029463 OMIM:259500 Orphanet:ORPHA668 SNOMED CT:21708004 MedGen:C2675080 |
| 27404 | G245S | Likely pathogenic;Pathogenic | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 MedGen:C0001418 MedGen:C0027672 SNOMED CT:699346009 MedGen:C0029463 OMIM:259500 Orphanet:ORPHA668 SNOMED CT:2170800 |
| 27405 | R273H | Pathogenic | Germline;somatic | GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 MedGen:C0027672 SNOMED CT:699346009 MedGen:C0238461 |
| 27407 | R280T | Pathogenic;Uncertain significance | Germline;somatic | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 MedGen:C2931822 OMIM:607107 Orphanet:ORPHA150 |
| 27408 | P151T | Likely pathogenic;Pathogenic | Germline;somatic | MedGen:C0027672 SNOMED CT:699346009 MedGen:C0858252 |
| 27409 | P151S | Pathogenic | Germline;somatic | MedGen:C0858252 MedGen:CN221809 |
| 27411 | L257Q | Pathogenic | Germline | MedGen:C2675080 |
| 27413 | R175H | Pathogenic | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 MedGen:C0027672 SNOMED CT:699346009 |
| 27422 | Y220S | Pathogenic | Germline | GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 |
| 27423 | E285V | Pathogenic | Germline | MedGen:C0431109 MedGen:C1859973 |
| 52763 | R273C | Likely pathogenic;Pathogenic;Uncertain significance | Germline;somatic | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 MedGen:C0027672 SNOMED CT:699346009 MedGen:C0376358 OMIM:176807 Orphanet:ORPHA1331 SNOMED CT:399068003 |
| 106675 | M246V | Likely pathogenic;Pathogenic | Germline;somatic | MedGen:C0027672 SNOMED CT:699346009 MedGen:CN221809 |
| 133267 | R156C | Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 133269 | R158C | Likely pathogenic;Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 133271 | Y163C | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 133276 | Y220C | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 133279 | M246R | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 150515 | C141Y | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 150535 | R282G | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 150812 | K120E | Pathogenic;Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 150815 | V122M | Uncertain significance | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 MedGen:C0027672 SNOMED CT:699346009 |
| 150855 | D281G | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 150938 | I232T | Likely pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 151595 | R249W | Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 151677 | R158H | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 151897 | Y236D | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 152145 | G108S | Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 152371 | V143M | Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 152405 | Y107C | Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 152428 | M237I | Likely pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 152480 | P152L | Pathogenic | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 MedGen:C0027672 SNOMED CT:699346009 |
| 176503 | S241C | Likely pathogenic | Germline | GeneReviews:NBK1311 MedGen:C1835398 OMIM:151623 Orphanet:ORPHA524 |
| 180995 | D281V | Pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 181000 | C238Y | Likely pathogenic;Uncertain significance | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 MedGen:C0027672 SNOMED CT:699346009 |
| 181001 | M237V | Pathogenic;Uncertain significance | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 MedGen:C0027672 SNOMED CT:699346009 |
| 181013 | A159P | Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 185350 | V272M | Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 185354 | N247I | Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 185371 | C176Y | Likely pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 185375 | Y163D | Likely pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 185387 | A138V | Uncertain significance | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 185394 | T125M | Likely pathogenic | Germline | MedGen:C0027672 SNOMED CT:699346009 |
| 213392 | C275Y | Likely pathogenic | Germline | GeneReviews:NBK1311 MedGen:C0085390 Orphanet:ORPHA524 SNOMED CT:428850001 |
| Top |
| Pharmacological information for TP53 |
 Gene expression profile of anticancer drug treated cell-lines (CCLE) Gene expression profile of anticancer drug treated cell-lines (CCLE)Heatmap showing the correlation between gene expression and drug response across all the cell-lines. We chose the top 20 among 138 drugs.We used Pearson's correlation coefficient. |
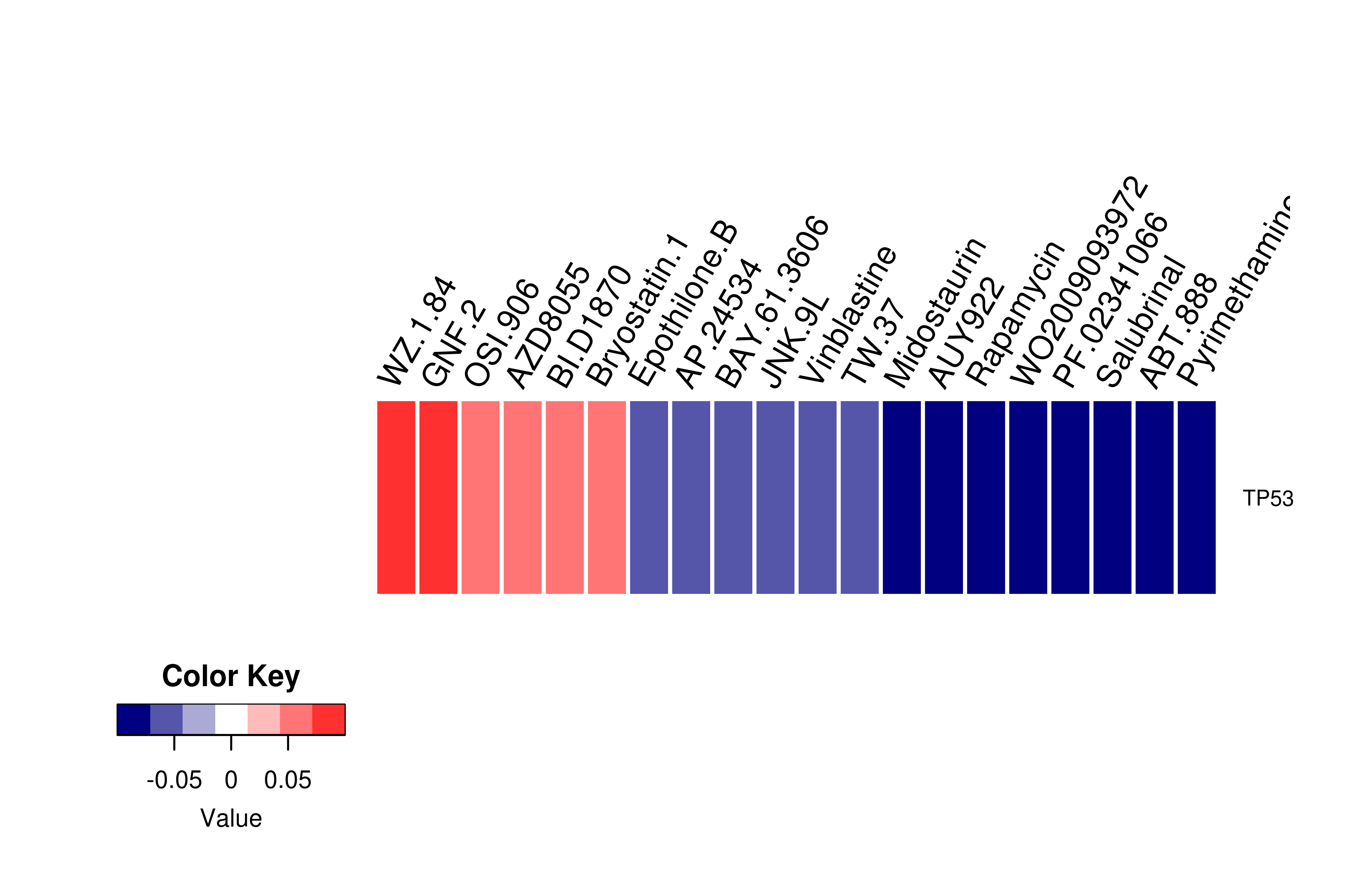 |
 Gene-centered drug-gene interaction network Gene-centered drug-gene interaction network |
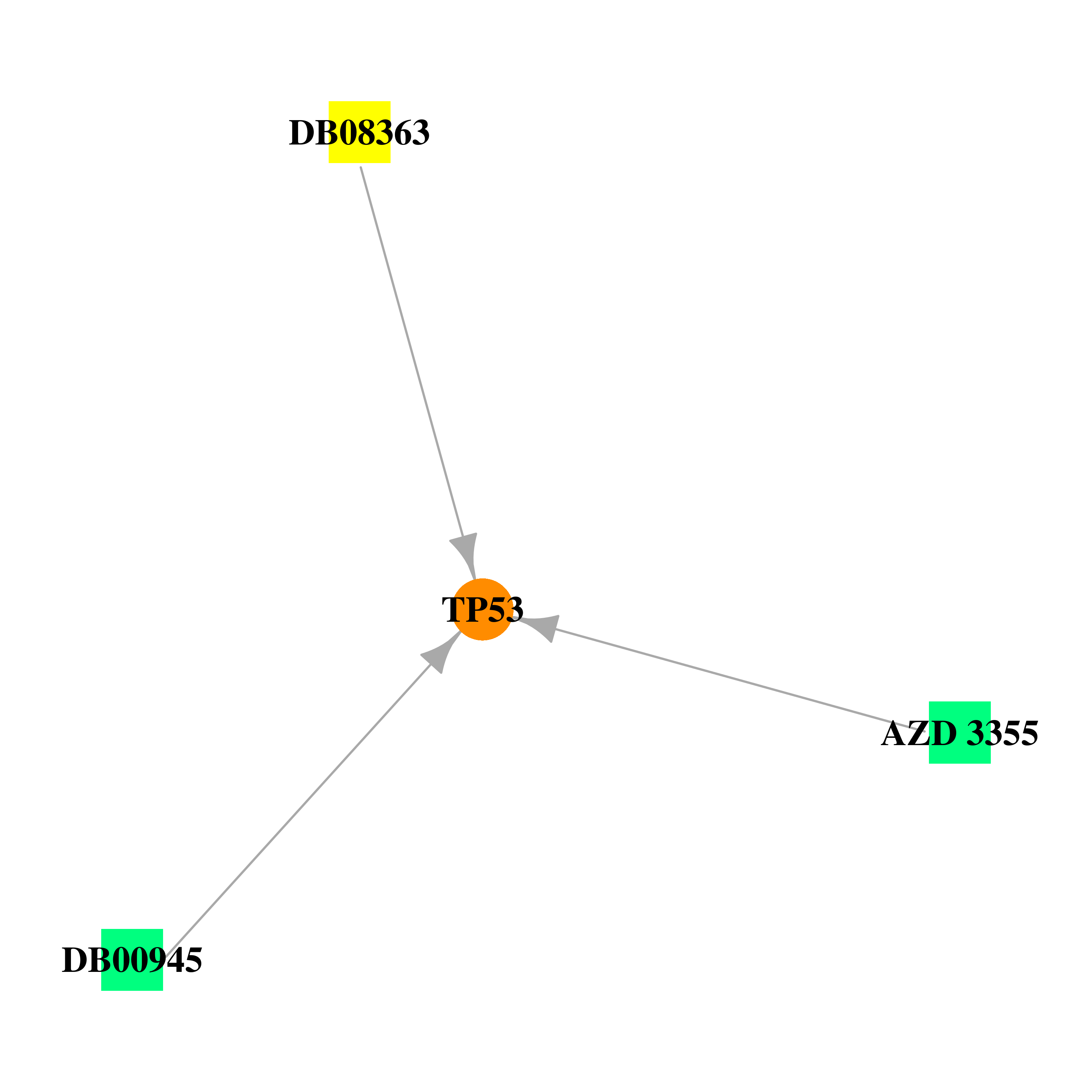 |
 Drug information targeting mutLBSgene (Approved drugs only) Drug information targeting mutLBSgene (Approved drugs only) |
| Drug status | DrugBank ID | Name | Type | Drug structure |
| Approved|vet_approved | DB00945 | Acetylsalicylic acid | Small molecule |  |
| Investigational | DB05404 | AZD 3355 | Small molecule |  |
| Experimental | DB08363 | 1-(9-ethyl-9H-carbazol-3-yl)-N-methylmethanamine | Small molecule | 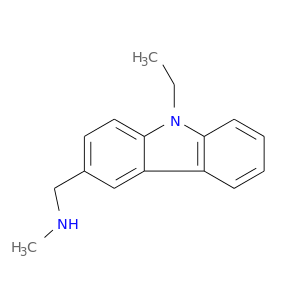 |
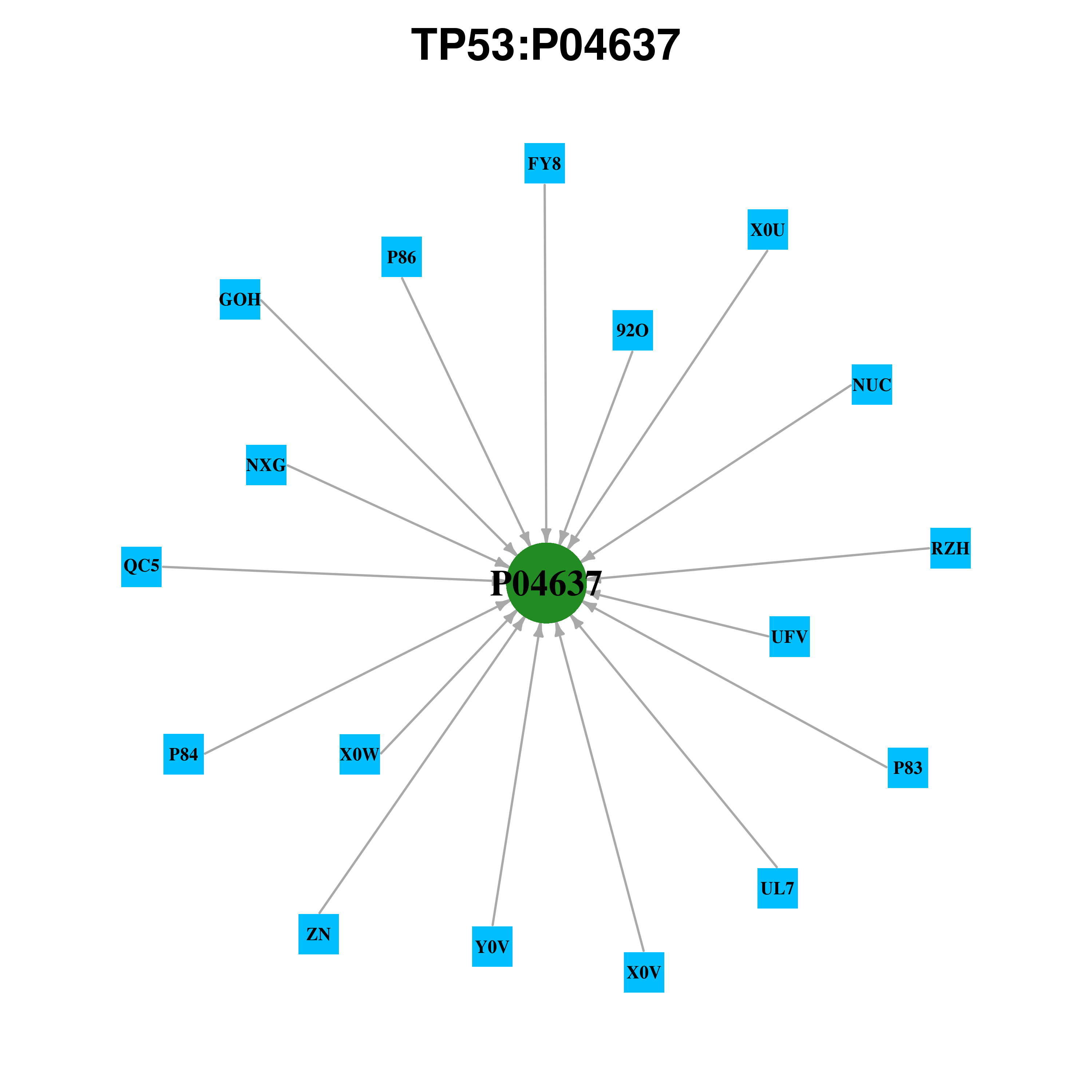
 Gene-centered ligand-gene interaction network Gene-centered ligand-gene interaction network |
 |
 Ligands binding to mutated ligand binding site of TP53 go to BioLip Ligands binding to mutated ligand binding site of TP53 go to BioLip |
| Ligand ID | Ligand short name | Ligand long name | PDB ID | PDB name | mutLBS | NUC | Nucleic Acids | 2ac0 | C | A119 K120 | NUC | Nucleic Acids | 2ata | D | A119 K120 | NUC | Nucleic Acids | 3d0a | D | A119 K120 | NUC | Nucleic Acids | 4hje | B | A119 K120 | NUC | Nucleic Acids | 4hje | D | A119 K120 | NUC | Nucleic Acids | 4ibu | B | A119 K120 M243 R248 R283 | NUC | Nucleic Acids | 3igk | A | A119 K120 Q167 R248 R280 | NUC | Nucleic Acids | 3igl | A | A119 K120 Q167 R248 R280 | NUC | Nucleic Acids | 2ac0 | A | A119 K120 R248 | NUC | Nucleic Acids | 2ac0 | D | A119 K120 R248 | NUC | Nucleic Acids | 2ady | A | A119 K120 R248 | NUC | Nucleic Acids | 2ahi | D | A119 K120 R248 | NUC | Nucleic Acids | 2ata | C | A119 K120 R248 | NUC | Nucleic Acids | 3d0a | B | A119 K120 R248 | NUC | Nucleic Acids | 3kmd | D | A119 K120 R248 | NUC | Nucleic Acids | 3q05 | B | A119 K120 R248 | NUC | Nucleic Acids | 4ibu | D | A119 K120 R248 | NUC | Nucleic Acids | 1tup | B | A119 K120 R248 R280 | NUC | Nucleic Acids | 1tsr | B | A119 K120 R248 R280 R283 | NUC | Nucleic Acids | 3d0a | A | A119 K120 V122 | NUC | Nucleic Acids | 4ibu | A | A119 K120 V122 | NUC | Nucleic Acids | 2ahi | A | A119 K120 V122 R248 | NUC | Nucleic Acids | 2ahi | C | A119 T123 | NUC | Nucleic Acids | 2ata | C | A119 T123 | NUC | Nucleic Acids | 2ac0 | C | A119 V122 T123 | ZN | ZINC(2+) | 1gzh | A | C176 C238 C242 | ZN | ZINC(2+) | 1gzh | C | C176 C238 C242 | ZN | ZINC(2+) | 1kzy | A | C176 C238 C242 | ZN | ZINC(2+) | 1kzy | B | C176 C238 C242 | ZN | ZINC(2+) | 1tsr | A | C176 C238 C242 | ZN | ZINC(2+) | 1tsr | B | C176 C238 C242 | ZN | ZINC(2+) | 1tsr | C | C176 C238 C242 | ZN | ZINC(2+) | 1tup | A | C176 C238 C242 | ZN | ZINC(2+) | 1tup | B | C176 C238 C242 | ZN | ZINC(2+) | 1tup | C | C176 C238 C242 | ZN | ZINC(2+) | 1uol | A | C176 C238 C242 | ZN | ZINC(2+) | 1uol | B | C176 C238 C242 | ZN | ZINC(2+) | 1ycs | A | C176 C238 C242 | ZN | ZINC(2+) | 2ac0 | A | C176 C238 C242 | ZN | ZINC(2+) | 2ac0 | B | C176 C238 C242 | ZN | ZINC(2+) | 2ac0 | C | C176 C238 C242 | ZN | ZINC(2+) | 2ac0 | D | C176 C238 C242 | ZN | ZINC(2+) | 2ady | A | C176 C238 C242 | ZN | ZINC(2+) | 2ady | B | C176 C238 C242 | ZN | ZINC(2+) | 2ahi | A | C176 C238 C242 | ZN | ZINC(2+) | 2ahi | B | C176 C238 C242 | ZN | ZINC(2+) | 2ahi | C | C176 C238 C242 | ZN | ZINC(2+) | 2ahi | D | C176 C238 C242 | ZN | ZINC(2+) | 2ata | A | C176 C238 C242 | ZN | ZINC(2+) | 2ata | B | C176 C238 C242 | ZN | ZINC(2+) | 2ata | C | C176 C238 C242 | ZN | ZINC(2+) | 2ata | D | C176 C238 C242 | ZN | ZINC(2+) | 2bim | A | C176 C238 C242 | ZN | ZINC(2+) | 2bim | B | C176 C238 C242 | ZN | ZINC(2+) | 2bin | A | C176 C238 C242 | ZN | ZINC(2+) | 2bio | A | C176 C238 C242 | ZN | ZINC(2+) | 2bip | A | C176 C238 C242 | ZN | ZINC(2+) | 2biq | A | C176 C238 C242 | ZN | ZINC(2+) | 2fej | A | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | M | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | N | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | O | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | P | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | Q | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | R | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | S | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | T | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | U | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | V | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | W | C176 C238 C242 | ZN | ZINC(2+) | 2h1l | X | C176 C238 C242 | ZN | ZINC(2+) | 2j1w | A | C176 C238 C242 | ZN | ZINC(2+) | 2j1w | B | C176 C238 C242 | ZN | ZINC(2+) | 2j1x | A | C176 C238 C242 | ZN | ZINC(2+) | 2j1x | B | C176 C238 C242 | ZN | ZINC(2+) | 2j1y | A | C176 C238 C242 | ZN | ZINC(2+) | 2j1y | B | C176 C238 C242 | ZN | ZINC(2+) | 2j1y | C | C176 C238 C242 | ZN | ZINC(2+) | 2j1y | D | C176 C238 C242 | ZN | ZINC(2+) | 2j1z | A | C176 C238 C242 | ZN | ZINC(2+) | 2j1z | B | C176 C238 C242 | ZN | ZINC(2+) | 2j20 | A | C176 C238 C242 | ZN | ZINC(2+) | 2j20 | B | C176 C238 C242 | ZN | ZINC(2+) | 2j21 | A | C176 C238 C242 | ZN | ZINC(2+) | 2j21 | B | C176 C238 C242 | ZN | ZINC(2+) | 2ocj | A | C176 C238 C242 | ZN | ZINC(2+) | 2ocj | B | C176 C238 C242 | ZN | ZINC(2+) | 2ocj | C | C176 C238 C242 | ZN | ZINC(2+) | 2ocj | D | C176 C238 C242 | ZN | ZINC(2+) | 2pcx | A | C176 C238 C242 | ZN | ZINC(2+) | 2qvq | A | C176 C238 C242 | ZN | ZINC(2+) | 2qxa | A | C176 C238 C242 | ZN | ZINC(2+) | 2qxa | B | C176 C238 C242 | ZN | ZINC(2+) | 2qxa | C | C176 C238 C242 | ZN | ZINC(2+) | 2qxa | D | C176 C238 C242 | ZN | ZINC(2+) | 2qxb | A | C176 C238 C242 | ZN | ZINC(2+) | 2qxb | B | C176 C238 C242 | ZN | ZINC(2+) | 2qxb | C | C176 C238 C242 | ZN | ZINC(2+) | 2qxb | D | C176 C238 C242 | ZN | ZINC(2+) | 2qxc | A | C176 C238 C242 | ZN | ZINC(2+) | 2qxc | B | C176 C238 C242 | ZN | ZINC(2+) | 2qxc | C | C176 C238 C242 | ZN | ZINC(2+) | 2qxc | D | C176 C238 C242 | ZN | ZINC(2+) | 2vuk | A | C176 C238 C242 | ZN | ZINC(2+) | 2vuk | B | C176 C238 C242 | ZN | ZINC(2+) | 2x0u | A | C176 C238 C242 | ZN | ZINC(2+) | 2x0u | B | C176 C238 C242 | ZN | ZINC(2+) | 2x0v | A | C176 C238 C242 | ZN | ZINC(2+) | 2x0v | B | C176 C238 C242 | ZN | ZINC(2+) | 2x0w | A | C176 C238 C242 | ZN | ZINC(2+) | 2x0w | B | C176 C238 C242 | ZN | ZINC(2+) | 2xwr | A | C176 C238 C242 | ZN | ZINC(2+) | 2xwr | B | C176 C238 C242 | ZN | ZINC(2+) | 2ybg | A | C176 C238 C242 | ZN | ZINC(2+) | 2ybg | B | C176 C238 C242 | ZN | ZINC(2+) | 2ybg | C | C176 C238 C242 | ZN | ZINC(2+) | 2ybg | D | C176 C238 C242 | ZN | ZINC(2+) | 3d05 | A | C176 C238 C242 | ZN | ZINC(2+) | 3d06 | A | C176 C238 C242 | ZN | ZINC(2+) | 3d07 | A | C176 C238 C242 | ZN | ZINC(2+) | 3d07 | B | C176 C238 C242 | ZN | ZINC(2+) | 3d08 | A | C176 C238 C242 | ZN | ZINC(2+) | 3d09 | A | C176 C238 C242 | ZN | ZINC(2+) | 3d0a | A | C176 C238 C242 | ZN | ZINC(2+) | 3d0a | B | C176 C238 C242 | ZN | ZINC(2+) | 3d0a | C | C176 C238 C242 | ZN | ZINC(2+) | 3d0a | D | C176 C238 C242 | ZN | ZINC(2+) | 3igk | A | C176 C238 C242 | ZN | ZINC(2+) | 3igl | A | C176 C238 C242 | ZN | ZINC(2+) | 3kmd | A | C176 C238 C242 | ZN | ZINC(2+) | 3kmd | B | C176 C238 C242 | ZN | ZINC(2+) | 3kmd | D | C176 C238 C242 | ZN | ZINC(2+) | 3kmd | C | C176 C238 C242 | ZN | ZINC(2+) | 3kz8 | A | C176 C238 C242 | ZN | ZINC(2+) | 3kz8 | B | C176 C238 C242 | ZN | ZINC(2+) | 3q01 | A | C176 C238 C242 | ZN | ZINC(2+) | 3q01 | B | C176 C238 C242 | ZN | ZINC(2+) | 3q05 | A | C176 C238 C242 | ZN | ZINC(2+) | 3q05 | B | C176 C238 C242 | ZN | ZINC(2+) | 3q05 | C | C176 C238 C242 | ZN | ZINC(2+) | 3q05 | D | C176 C238 C242 | ZN | ZINC(2+) | 3q06 | A | C176 C238 C242 | ZN | ZINC(2+) | 3q06 | B | C176 C238 C242 | ZN | ZINC(2+) | 3q06 | C | C176 C238 C242 | ZN | ZINC(2+) | 3q06 | D | C176 C238 C242 | ZN | ZINC(2+) | 3ts8 | A | C176 C238 C242 | ZN | ZINC(2+) | 3ts8 | B | C176 C238 C242 | ZN | ZINC(2+) | 3ts8 | C | C176 C238 C242 | ZN | ZINC(2+) | 3ts8 | D | C176 C238 C242 | ZN | ZINC(2+) | 4agl | A | C176 C238 C242 | ZN | ZINC(2+) | 4agl | B | C176 C238 C242 | ZN | ZINC(2+) | 4agm | A | C176 C238 C242 | ZN | ZINC(2+) | 4agm | B | C176 C238 C242 | ZN | ZINC(2+) | 4agn | A | C176 C238 C242 | ZN | ZINC(2+) | 4agn | B | C176 C238 C242 | ZN | ZINC(2+) | 4ago | A | C176 C238 C242 | ZN | ZINC(2+) | 4ago | B | C176 C238 C242 | ZN | ZINC(2+) | 4agp | A | C176 C238 C242 | ZN | ZINC(2+) | 4agp | B | C176 C238 C242 | ZN | ZINC(2+) | 4agq | A | C176 C238 C242 | ZN | ZINC(2+) | 4agq | B | C176 C238 C242 | ZN | ZINC(2+) | 3zme | A | C176 C238 C242 | ZN | ZINC(2+) | 3zme | B | C176 C238 C242 | ZN | ZINC(2+) | 4hje | A | C176 C238 C242 | ZN | ZINC(2+) | 4hje | B | C176 C238 C242 | ZN | ZINC(2+) | 4hje | C | C176 C238 C242 | ZN | ZINC(2+) | 4hje | D | C176 C238 C242 | ZN | ZINC(2+) | 4kvp | A | C176 C238 C242 | ZN | ZINC(2+) | 4kvp | B | C176 C238 C242 | ZN | ZINC(2+) | 4kvp | C | C176 C238 C242 | ZN | ZINC(2+) | 4kvp | D | C176 C238 C242 | ZN | ZINC(2+) | 4lo9 | A | C176 C238 C242 | ZN | ZINC(2+) | 4lo9 | B | C176 C238 C242 | ZN | ZINC(2+) | 4lo9 | C | C176 C238 C242 | ZN | ZINC(2+) | 4lo9 | D | C176 C238 C242 | ZN | ZINC(2+) | 4loe | A | C176 C238 C242 | ZN | ZINC(2+) | 4loe | B | C176 C238 C242 | ZN | ZINC(2+) | 4loe | C | C176 C238 C242 | ZN | ZINC(2+) | 4loe | D | C176 C238 C242 | ZN | ZINC(2+) | 4lof | A | C176 C238 C242 | ZN | ZINC(2+) | 4ibq | A | C176 C238 C242 | ZN | ZINC(2+) | 4ibq | B | C176 C238 C242 | ZN | ZINC(2+) | 4ibq | C | C176 C238 C242 | ZN | ZINC(2+) | 4ibq | D | C176 C238 C242 | ZN | ZINC(2+) | 4ibs | A | C176 C238 C242 | ZN | ZINC(2+) | 4ibs | B | C176 C238 C242 | ZN | ZINC(2+) | 4ibs | C | C176 C238 C242 | ZN | ZINC(2+) | 4ibs | D | C176 C238 C242 | ZN | ZINC(2+) | 4ibt | A | C176 C238 C242 | ZN | ZINC(2+) | 4ibt | B | C176 C238 C242 | ZN | ZINC(2+) | 4ibt | C | C176 C238 C242 | ZN | ZINC(2+) | 4ibt | D | C176 C238 C242 | ZN | ZINC(2+) | 4ibu | A | C176 C238 C242 | ZN | ZINC(2+) | 4ibu | B | C176 C238 C242 | ZN | ZINC(2+) | 4ibu | C | C176 C238 C242 | ZN | ZINC(2+) | 4ibu | D | C176 C238 C242 | ZN | ZINC(2+) | 4ibv | A | C176 C238 C242 | ZN | ZINC(2+) | 4ibw | A | C176 C238 C242 | ZN | ZINC(2+) | 4iby | A | C176 C238 C242 | ZN | ZINC(2+) | 4iby | B | C176 C238 C242 | ZN | ZINC(2+) | 4ibz | A | C176 C238 C242 | ZN | ZINC(2+) | 4ibz | B | C176 C238 C242 | ZN | ZINC(2+) | 4ibz | C | C176 C238 C242 | ZN | ZINC(2+) | 4ibz | D | C176 C238 C242 | ZN | ZINC(2+) | 4ijt | A | C176 C238 C242 | ZN | ZINC(2+) | 4mzi | A | C176 C238 C242 | ZN | ZINC(2+) | 4mzr | A | C176 C238 C242 | ZN | ZINC(2+) | 4mzr | B | C176 C238 C242 | ZN | ZINC(2+) | 4mzr | C | C176 C238 C242 | ZN | ZINC(2+) | 4mzr | D | C176 C238 C242 | ZN | ZINC(2+) | 2mej | B | C176 C238 C242 | ZN | ZINC(2+) | 4qo1 | B | C176 C238 C242 | ZN | ZINC(2+) | 5a7b | A | C176 C238 C242 | ZN | ZINC(2+) | 5a7b | B | C176 C238 C242 | ZN | ZINC(2+) | 5ab9 | A | C176 C238 C242 | ZN | ZINC(2+) | 5ab9 | B | C176 C238 C242 | ZN | ZINC(2+) | 5aba | A | C176 C238 C242 | ZN | ZINC(2+) | 5aba | B | C176 C238 C242 | ZN | ZINC(2+) | 5aoi | A | C176 C238 C242 | ZN | ZINC(2+) | 5aoi | B | C176 C238 C242 | ZN | ZINC(2+) | 5aoj | A | C176 C238 C242 | ZN | ZINC(2+) | 5aoj | B | C176 C238 C242 | ZN | ZINC(2+) | 5aok | A | C176 C238 C242 | ZN | ZINC(2+) | 5aok | B | C176 C238 C242 | ZN | ZINC(2+) | 5aol | A | C176 C238 C242 | ZN | ZINC(2+) | 5aol | B | C176 C238 C242 | ZN | ZINC(2+) | 5aom | A | C176 C238 C242 | ZN | ZINC(2+) | 5aom | B | C176 C238 C242 | ZN | ZINC(2+) | 5ecg | A | C176 C238 C242 | ZN | ZINC(2+) | 5ecg | B | C176 C238 C242 | UFV | 3-BROMO-5-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE | 5aol | A | F109 L145 V157 E221 T230 L257 | NUC | Nucleic Acids | 1tsr | C | K120 | NUC | Nucleic Acids | 1tup | C | K120 | NUC | Nucleic Acids | 3d0a | C | K120 | NUC | Nucleic Acids | 3q06 | B | K120 | NUC | Nucleic Acids | 3q06 | C | K120 | NUC | Nucleic Acids | 4ibu | C | K120 | NUC | Nucleic Acids | 4mzr | B | K120 | NUC | Nucleic Acids | 2ac0 | C | K120 N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ahi | D | K120 N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ata | B | K120 N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ahi | C | K120 N239 S241 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ahi | B | K120 R248 | NUC | Nucleic Acids | 2ahi | C | K120 R248 | NUC | Nucleic Acids | 2ata | A | K120 R248 | NUC | Nucleic Acids | 3kmd | A | K120 R248 | NUC | Nucleic Acids | 3kmd | B | K120 R248 | NUC | Nucleic Acids | 3kmd | C | K120 R248 | NUC | Nucleic Acids | 3q05 | C | K120 R248 | NUC | Nucleic Acids | 3ts8 | B | K120 R248 | NUC | Nucleic Acids | 3ts8 | C | K120 R248 | NUC | Nucleic Acids | 4hje | A | K120 R248 | NUC | Nucleic Acids | 4hje | C | K120 R248 | NUC | Nucleic Acids | 4mzr | C | K120 R248 | NUC | Nucleic Acids | 3kz8 | A | K120 R248 R280 | NUC | Nucleic Acids | 3kz8 | B | K120 R248 R280 | NUC | Nucleic Acids | 2ady | A | K120 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ady | B | K120 S241 R248 R273 C275 A276 C277 R280 | UL7 | 1-{1-[(3-BROMO-5-CHLORO-2-HYDROXYPHENYL)METHYL] PIPERIDIN-4-YL}PIPERIDIN-4-OL | 5aba | A | L145 D148 T150 E221 P223 T230 | P86 | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4,6-DIIODOPHENOL | 4agm | A | L145 D148 T150 P151 E221 P223 T230 | UL7 | 1-{1-[(3-BROMO-5-CHLORO-2-HYDROXYPHENYL)METHYL] PIPERIDIN-4-YL}PIPERIDIN-4-OL | 5aba | B | L145 D148 T150 P151 E221 P223 T230 | FY8 | N-(5-CHLORANYL-2-OXIDANYL-PHENYL)PIPERIDINE- 4-CARBOXAMIDE | 5aom | A | L145 D148 T150 P151 E221 P223 T230 | FY8 | N-(5-CHLORANYL-2-OXIDANYL-PHENYL)PIPERIDINE- 4-CARBOXAMIDE | 5aom | B | L145 D148 T150 P151 E221 P223 T230 | KMN | 2-[[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL]-6-ETHYNYL- 4-(3-PHENOXYPROP-1-YNYL)PHENOL | 5a7b | B | L145 D148 T150 P151 P152 E221 P223 | P74 | TERT-BUTYL [3-(3-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-HYDROXY-5-IODOPHENYL)PROP-2-YN-1-YL]CARBAMATE | 4ago | A | L145 D148 T150 P151 P152 E221 P223 T230 | P51 | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-6-IODO-4-(3-PHENOXYPROP-1-YN-1-YL)PHENOL | 4agp | A | L145 D148 T150 P151 P152 E221 P223 T230 | P96 | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-6-IODO-4-[3-(PHENYLAMINO)PROP-1-YN-1-YL]PHENOL | 4agq | A | L145 D148 T150 P151 P152 E221 P223 T230 | KMN | 2-[[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL]-6-ETHYNYL- 4-(3-PHENOXYPROP-1-YNYL)PHENOL | 5a7b | A | L145 D148 T150 P151 P152 E221 P223 T230 | NXG | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-(3-HYDROXYPROP-1-YN-1-YL)-6-IODOPHENOL | 4agn | A | L145 D148 T150 P151 P152 P153 E221 P223 T230 | NXG | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-(3-HYDROXYPROP-1-YN-1-YL)-6-IODOPHENOL | 4agn | B | L145 D148 T150 P151 P152 P153 E221 P223 T230 | P74 | TERT-BUTYL [3-(3-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-HYDROXY-5-IODOPHENYL)PROP-2-YN-1-YL]CARBAMATE | 4ago | B | L145 D148 T150 P151 P152 P153 E221 P223 T230 | P51 | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-6-IODO-4-(3-PHENOXYPROP-1-YN-1-YL)PHENOL | 4agp | B | L145 D148 T150 P151 P152 P153 E221 P223 T230 | P96 | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-6-IODO-4-[3-(PHENYLAMINO)PROP-1-YN-1-YL]PHENOL | 4agq | B | L145 D148 T150 P151 P152 P153 E221 P223 T230 | QC5 | 2-(4-(4-FLUOROPHENYL)-5-(1H-PYRROL-1-YL)-1H- PYRAZOL-1-YL)-N,N-DIMETHYLETHANAMINE | 3zme | A | L145 D148 T150 P151 P153 E221 P223 T230 | 92O | 7-ETHYL-3-(PIPERIDIN-4-YL)-1H-INDOLE | 5ab9 | A | L145 D148 T150 P223 D228 T230 | P83 | 1-(9-ETHYL-9H-CARBAZOL-3-YL)-N-METHYLMETHANAMINE | 2vuk | B | L145 T150 P151 E221 P223 D228 T230 | X0V | 3,4-DIAMINOBENZOTRIFLUORIDE | 2x0v | B | L145 T150 P151 E221 P223 T230 | P84 | 2,4-BIS(IODANYL)-6-[[METHYL-(1-METHYLPIPERIDIN-4-YL)AMINO]METHYL}PHENOL | 4agl | A | L145 T150 P151 E221 P223 T230 | P84 | 2,4-BIS(IODANYL)-6-[[METHYL-(1-METHYLPIPERIDIN-4-YL)AMINO]METHYL}PHENOL | 4agl | B | L145 T150 P151 E221 P223 T230 | GOH | 5-[2-CYCLOPROPYL-5-(1H-PYRROL-1-YL)-1,3- OXAZOL-4-YL]-1H-1,2,3,4-TETRAZOLE | 5aok | A | L145 T150 P151 P152 E221 P223 T230 | GOH | 5-[2-CYCLOPROPYL-5-(1H-PYRROL-1-YL)-1,3- OXAZOL-4-YL]-1H-1,2,3,4-TETRAZOLE | 5aok | B | L145 T150 P151 P152 E221 P223 T230 | X0U | 6,7-DIHYDRO-[1,4]DIOXINO[2,3-F][1,3]BENZOTHIAZOL-2-AMINE | 2x0u | B | L145 T150 P151 P152 T155 P223 T230 | QC5 | 2-(4-(4-FLUOROPHENYL)-5-(1H-PYRROL-1-YL)-1H- PYRAZOL-1-YL)-N,N-DIMETHYLETHANAMINE | 3zme | B | L145 T150 P151 P153 E221 P223 T230 | 92O | 7-ETHYL-3-(PIPERIDIN-4-YL)-1H-INDOLE | 5ab9 | B | L145 T150 P151 P223 D228 T230 | P86 | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4,6-DIIODOPHENOL | 4agm | B | L145 W146 D148 T150 P151 E221 P223 T230 | Y0V | 2-HYDROXY-3,5-DIIODO-4-(1H-PYRROL-1-YL) BENZOIC ACID | 5aoj | A | L145 W146 T150 P151 E221 P223 T230 | Y0V | 2-HYDROXY-3,5-DIIODO-4-(1H-PYRROL-1-YL) BENZOIC ACID | 5aoj | B | L145 W146 T150 P151 E221 P223 T230 | NUC | Nucleic Acids | 4ibw | A | N239 S241 R248 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ac0 | A | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ac0 | D | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ata | C | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ata | D | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3d0a | C | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3d0a | D | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3kmd | A | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3kmd | B | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3kmd | D | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3kmd | C | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 4hje | A | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 4hje | C | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 4hje | D | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 4mzr | B | N239 S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ahi | A | N239 S241 R248 R273 C275 A276 R280 | FY8 | N-(5-CHLORANYL-2-OXIDANYL-PHENYL)PIPERIDINE- 4-CARBOXAMIDE | 5aom | B | P151 P152 P153 T155 | FY8 | N-(5-CHLORANYL-2-OXIDANYL-PHENYL)PIPERIDINE- 4-CARBOXAMIDE | 5aom | A | P151 P152 P153 T155 E221 | NUC | Nucleic Acids | 1tsr | A | Q165 Q167 H168 | NUC | Nucleic Acids | 1tup | A | Q165 Q167 H168 | NUC | Nucleic Acids | 4mzr | A | R248 R283 | NUC | Nucleic Acids | 3q06 | C | S241 A276 C277 R280 | NUC | Nucleic Acids | 4ibu | B | S241 C275 A276 C277 R280 | NUC | Nucleic Acids | 4ibu | C | S241 C275 A276 C277 R280 | NUC | Nucleic Acids | 4ibu | D | S241 C275 A276 C277 R280 | NUC | Nucleic Acids | 4ibv | A | S241 C275 A276 C277 R280 | NUC | Nucleic Acids | 1tsr | C | S241 R248 | NUC | Nucleic Acids | 1tup | C | S241 R248 | NUC | Nucleic Acids | 4ibu | A | S241 R248 C275 A276 C277 R280 | NUC | Nucleic Acids | 1tsr | B | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 1tup | B | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ac0 | B | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 2ahi | B | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3q05 | B | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3q06 | B | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3ts8 | A | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3ts8 | D | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 4hje | B | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 4mzr | D | S241 R248 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3d0a | A | S241 R248 R273 C275 A276 R280 | NUC | Nucleic Acids | 3q05 | A | S241 R248 R273 C275 A276 R280 | NUC | Nucleic Acids | 3q05 | C | S241 R248 R273 C275 A276 R280 | NUC | Nucleic Acids | 3ts8 | C | S241 R248 R273 C275 A276 R280 | NUC | Nucleic Acids | 4mzr | C | S241 R273 A276 C277 R280 | NUC | Nucleic Acids | 2ata | A | S241 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3d0a | B | S241 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3q05 | D | S241 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3q06 | A | S241 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3q06 | D | S241 R273 C275 A276 C277 R280 | NUC | Nucleic Acids | 3ts8 | B | S241 R273 C275 A276 R280 | NUC | Nucleic Acids | 4mzr | A | S241 R273 C275 A276 R280 | NUC | Nucleic Acids | 2ac0 | B | T118 A119 K120 R248 G279 R280 R283 | NUC | Nucleic Acids | 2ady | B | T118 A119 K120 R248 R283 | NUC | Nucleic Acids | 2ahi | A | T123 K139 | RZH | 2-(5-BROMO-7-ETHYL-2-METHYL-1H-INDOLE-3-YL) ETHAN-1-AMIN | 5aoi | B | T150 P151 P152 E221 P223 | 92O | 7-ETHYL-3-(PIPERIDIN-4-YL)-1H-INDOLE | 5ab9 | A | T150 P151 P152 P153 | 92O | 7-ETHYL-3-(PIPERIDIN-4-YL)-1H-INDOLE | 5ab9 | B | T150 P151 P152 P153 | X0V | 3,4-DIAMINOBENZOTRIFLUORIDE | 2x0v | B | T150 P151 P152 P153 T155 E221 | X0W | 5,6-DIMETHOXY-2-METHYL-BENZOTHIAZOLE | 2x0w | B | T150 P151 P223 | NUC | Nucleic Acids | 3q05 | A | V122 R248 R283 | NUC | Nucleic Acids | 3q06 | A | V122 R248 R283 | NUC | Nucleic Acids | 3q05 | D | V122 R283 | NUC | Nucleic Acids | 2ac0 | A | V122 T123 | NUC | Nucleic Acids | 2ata | A | V122 T123 |
| Top |
| Conservation information for LBS of TP53 |
 Multiple alignments for P04637 in multiple species Multiple alignments for P04637 in multiple species |
| LBS | AA sequence | # species | Species | A119 | LHSGTAKSVTC | 2 | Homo sapiens, Macaca mulatta | A119 | LQSGTAKSVMC | 2 | Mus musculus, Rattus norvegicus | A119 | LQSGTAKSVTC | 1 | Bos taurus | A119 | LHSGTAKSVTW | 1 | Canis lupus familiaris | A276 | EVRVCACPGRD | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | C176 | EVVRRCPHHER | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | C229 | EVGSDCTTIHY | 2 | Homo sapiens, Macaca mulatta | C229 | EVGSDYTTIHY | 2 | Rattus norvegicus, Canis lupus familiaris | C229 | EAGSEYTTIHY | 1 | Mus musculus | C229 | EIDSECTTIHY | 1 | Bos taurus | C238 | HYNYMCNSSCM | 3 | Homo sapiens, Canis lupus familiaris, Macaca mulatta | C238 | HYKYMCNSSCM | 2 | Mus musculus, Rattus norvegicus | C238 | HYNFMCNSSCM | 1 | Bos taurus | C242 | MCNSSCMGGMN | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | C275 | FEVRVCACPGR | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | C277 | VRVCACPGRDR | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | D148 | VQLWVDSTPPP | 2 | Homo sapiens, Macaca mulatta | D148 | VQLWVSATPPA | 1 | Mus musculus | D148 | VQLWVTSTPPP | 1 | Rattus norvegicus | D148 | VQLWVDSPPPP | 1 | Bos taurus | D148 | VQLWVSSPPPP | 1 | Canis lupus familiaris | D228 | PEVGSDCTTIH | 2 | Homo sapiens, Macaca mulatta | D228 | PEVGSDYTTIH | 2 | Rattus norvegicus, Canis lupus familiaris | D228 | PEAGSEYTTIH | 1 | Mus musculus | D228 | PEIDSECTTIH | 1 | Bos taurus | E221 | VVVPYEPPEVG | 4 | Homo sapiens, Rattus norvegicus, Canis lupus familiaris, Macaca mulatta | E221 | VVVPYEPPEAG | 1 | Mus musculus | E221 | VVVPYESPEID | 1 | Bos taurus | F109 | QGNYGFHLGFL | 2 | Mus musculus, Rattus norvegicus | F109 | QGSYGFRLGFL | 1 | Homo sapiens | F109 | PGNYGFRLGFL | 1 | Bos taurus | F109 | PGTYGFRLGFL | 1 | Canis lupus familiaris | F109 | HGSYGFRLGFL | 1 | Macaca mulatta | G279 | VCACPGRDRRT | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | H168 | YKQSQHMTEVV | 2 | Homo sapiens, Macaca mulatta | H168 | YKKSQHMTEVV | 2 | Mus musculus, Rattus norvegicus | H168 | YKKLEHMTEVV | 1 | Bos taurus | H168 | YKKSEFVTEVV | 1 | Canis lupus familiaris | K120 | HSGTAKSVTCT | 2 | Homo sapiens, Macaca mulatta | K120 | QSGTAKSVMCT | 2 | Mus musculus, Rattus norvegicus | K120 | QSGTAKSVTCT | 1 | Bos taurus | K120 | HSGTAKSVTWT | 1 | Canis lupus familiaris | K139 | FCQLAKTCPVQ | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | L145 | TCPVQLWVDST | 2 | Homo sapiens, Macaca mulatta | L145 | TCPVQLWVSAT | 1 | Mus musculus | L145 | TCPVQLWVTST | 1 | Rattus norvegicus | L145 | TCPVQLWVDSP | 1 | Bos taurus | L145 | TCPVQLWVSSP | 1 | Canis lupus familiaris | L257 | LTIITLEDSSG | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Canis lupus familiaris, Macaca mulatta | L257 | LTIITLEDSCG | 1 | Bos taurus | M243 | CNSSCMGGMNR | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | N239 | YNYMCNSSCMG | 3 | Homo sapiens, Canis lupus familiaris, Macaca mulatta | N239 | YKYMCNSSCMG | 2 | Mus musculus, Rattus norvegicus | N239 | YNFMCNSSCMG | 1 | Bos taurus | P151 | WVDSTPPPGTR | 1 | Homo sapiens | P151 | WVSATPPAGSR | 1 | Mus musculus | P151 | WVTSTPPPGTR | 1 | Rattus norvegicus | P151 | WVDSPPPPGTR | 1 | Bos taurus | P151 | WVSSPPPPNTC | 1 | Canis lupus familiaris | P151 | WVDSTPPPGSR | 1 | Macaca mulatta | P152 | VDSTPPPGTRV | 1 | Homo sapiens | P152 | VSATPPAGSRV | 1 | Mus musculus | P152 | VTSTPPPGTRV | 1 | Rattus norvegicus | P152 | VDSPPPPGTRV | 1 | Bos taurus | P152 | VSSPPPPNTCV | 1 | Canis lupus familiaris | P152 | VDSTPPPGSRV | 1 | Macaca mulatta | P153 | DSTPPPGTRVR | 1 | Homo sapiens | P153 | SATPPAGSRVR | 1 | Mus musculus | P153 | TSTPPPGTRVR | 1 | Rattus norvegicus | P153 | DSPPPPGTRVR | 1 | Bos taurus | P153 | SSPPPPNTCVR | 1 | Canis lupus familiaris | P153 | DSTPPPGSRVR | 1 | Macaca mulatta | P222 | VVPYEPPEVGS | 4 | Homo sapiens, Rattus norvegicus, Canis lupus familiaris, Macaca mulatta | P222 | VVPYEPPEAGS | 1 | Mus musculus | P222 | VVPYESPEIDS | 1 | Bos taurus | P223 | VPYEPPEVGSD | 4 | Homo sapiens, Rattus norvegicus, Canis lupus familiaris, Macaca mulatta | P223 | VPYEPPEAGSE | 1 | Mus musculus | P223 | VPYESPEIDSE | 1 | Bos taurus | Q165 | MAIYKQSQHMT | 2 | Homo sapiens, Macaca mulatta | Q165 | MAIYKKSQHMT | 2 | Mus musculus, Rattus norvegicus | Q165 | MAIYKKLEHMT | 1 | Bos taurus | Q165 | MAIYKKSEFVT | 1 | Canis lupus familiaris | Q167 | IYKQSQHMTEV | 2 | Homo sapiens, Macaca mulatta | Q167 | IYKKSQHMTEV | 2 | Mus musculus, Rattus norvegicus | Q167 | IYKKLEHMTEV | 1 | Bos taurus | Q167 | IYKKSEFVTEV | 1 | Canis lupus familiaris | R248 | MGGMNRRPILT | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | R273 | NSFEVRVCACP | 4 | Homo sapiens, Bos taurus, Canis lupus familiaris, Macaca mulatta | R273 | DSFEVRVCACP | 2 | Mus musculus, Rattus norvegicus | R280 | CACPGRDRRTE | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | R283 | PGRDRRTEEEN | 6 | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris, Macaca mulatta | S121 | SGTAKSVTCTY | 3 | Homo sapiens, Bos taurus, Macaca mulatta | S121 | SGTAKSVMCTY | 2 | Mus musculus, Rattus norvegicus | S121 | SGTAKSVTWTY | 1 | Canis lupus familiaris | S149 | QLWVDSTPPPG | 2 | Homo sapiens, Macaca mulatta | S149 | QLWVSATPPAG | 1 | Mus musculus | S149 | QLWVTSTPPPG | 1 | Rattus norvegicus | S149 | QLWVDSPPPPG | 1 | Bos taurus | S149 | QLWVSSPPPPN | 1 | Canis lupus familiaris | S241 | YMCNSSCMGGM | 5 | Homo sapiens, Mus musculus, Rattus norvegicus, Canis lupus familiaris, Macaca mulatta | S241 | FMCNSSCMGGM | 1 | Bos taurus | T118 | FLHSGTAKSVT | 3 | Homo sapiens, Canis lupus familiaris, Macaca mulatta | T118 | FLQSGTAKSVM | 2 | Mus musculus, Rattus norvegicus | T118 | FLQSGTAKSVT | 1 | Bos taurus | T123 | TAKSVTCTYSP | 3 | Homo sapiens, Bos taurus, Macaca mulatta | T123 | TAKSVMCTYSP | 1 | Mus musculus | T123 | TAKSVMCTYSI | 1 | Rattus norvegicus | T123 | TAKSVTWTYSP | 1 | Canis lupus familiaris | T150 | LWVDSTPPPGT | 1 | Homo sapiens | T150 | LWVSATPPAGS | 1 | Mus musculus | T150 | LWVTSTPPPGT | 1 | Rattus norvegicus | T150 | LWVDSPPPPGT | 1 | Bos taurus | T150 | LWVSSPPPPNT | 1 | Canis lupus familiaris | T150 | LWVDSTPPPGS | 1 | Macaca mulatta | T155 | TPPPGTRVRAM | 2 | Homo sapiens, Rattus norvegicus | T155 | TPPAGSRVRAM | 1 | Mus musculus | T155 | PPPPGTRVRAM | 1 | Bos taurus | T155 | PPPPNTCVRAM | 1 | Canis lupus familiaris | T155 | TPPPGSRVRAM | 1 | Macaca mulatta | T230 | VGSDCTTIHYN | 2 | Homo sapiens, Macaca mulatta | T230 | AGSEYTTIHYK | 1 | Mus musculus | T230 | VGSDYTTIHYK | 1 | Rattus norvegicus | T230 | IDSECTTIHYN | 1 | Bos taurus | T230 | VGSDYTTIHYN | 1 | Canis lupus familiaris | V122 | GTAKSVTCTYS | 3 | Homo sapiens, Bos taurus, Macaca mulatta | V122 | GTAKSVMCTYS | 2 | Mus musculus, Rattus norvegicus | V122 | GTAKSVTWTYS | 1 | Canis lupus familiaris | V147 | PVQLWVDSTPP | 2 | Homo sapiens, Macaca mulatta | V147 | PVQLWVSATPP | 1 | Mus musculus | V147 | PVQLWVTSTPP | 1 | Rattus norvegicus | V147 | PVQLWVDSPPP | 1 | Bos taurus | V147 | PVQLWVSSPPP | 1 | Canis lupus familiaris | V157 | PPGTRVRAMAI | 3 | Homo sapiens, Rattus norvegicus, Bos taurus | V157 | PAGSRVRAMAI | 1 | Mus musculus | V157 | PPNTCVRAMAI | 1 | Canis lupus familiaris | V157 | PPGSRVRAMAI | 1 | Macaca mulatta | W146 | CPVQLWVDSTP | 2 | Homo sapiens, Macaca mulatta | W146 | CPVQLWVSATP | 1 | Mus musculus | W146 | CPVQLWVTSTP | 1 | Rattus norvegicus | W146 | CPVQLWVDSPP | 1 | Bos taurus | W146 | CPVQLWVSSPP | 1 | Canis lupus familiaris |
 |
Copyright © 2016-Present - The University of Texas Health Science Center at Houston |