Introduction
WebCSEA (Web-based Cell-type Specific Enrichment Analysis of Genes) provides a gene set query among a systematic collection of tissue-cell-type expression signatures. For each query, we will generate cell-type-specific enrichment analysis (CSEA) raw p-value, combined p-value by permutation-based method, genes shared between queried gene list and tissue-cell-type signatures, heatmap that interactively displays the cell-type specificity across 1,355 human tissue-cell types (TCs), summary of cell-type specificity by human organ system and top-ranked tissues and general cell types. Users can also filter, prioritize, and compare the CSEA results from cell types from one or multiple tissues. Users can further download all the results of CSEA and visualize them. Overall, WebCSEA provides a comprehensive map of major human TCs and uses a permutation-based method to overcome the bias raised from different lengths of signature genes among TCs as well as the length of query genes.
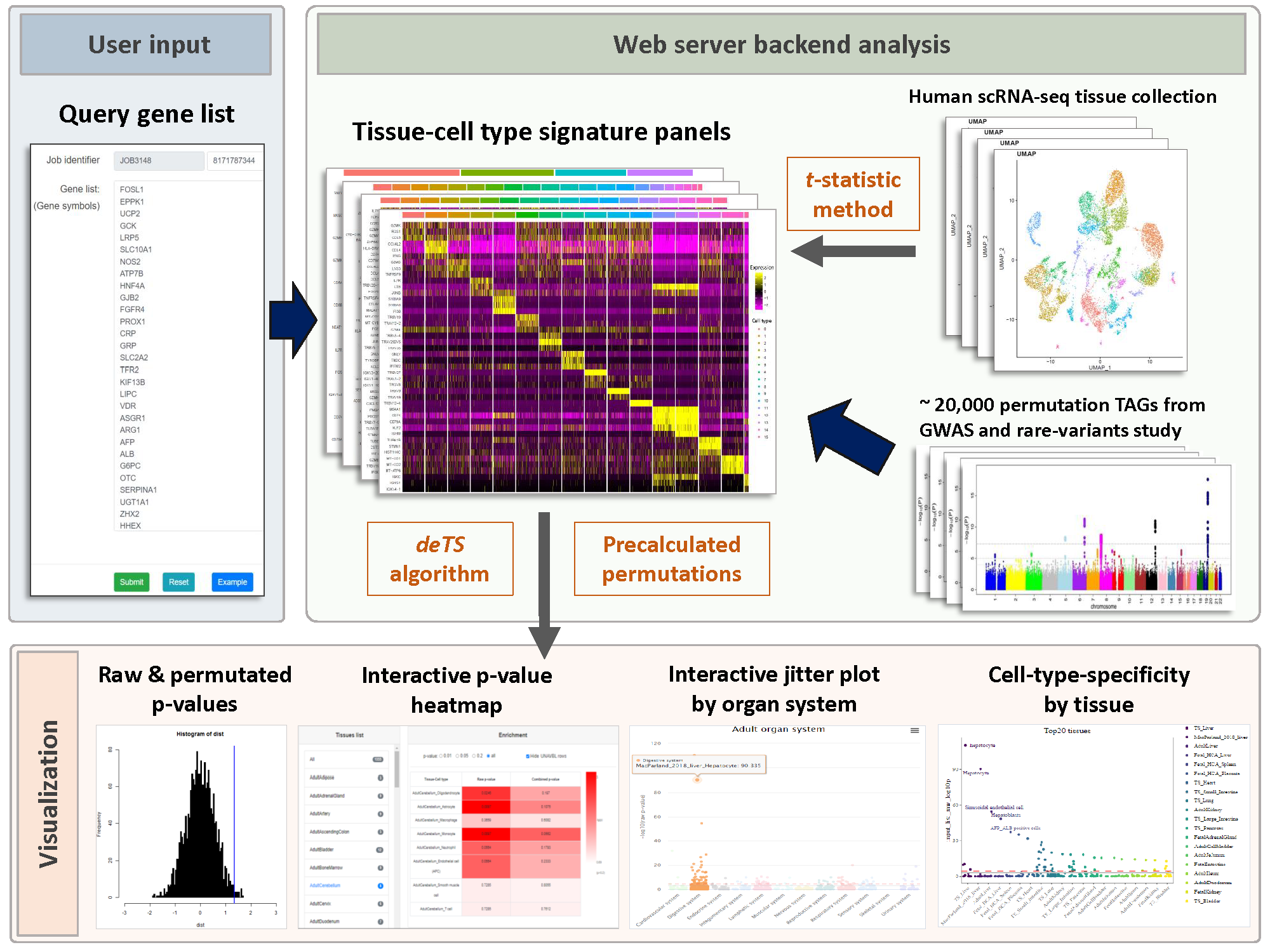
Data collection for constructing our tissue-cell type signature panel and permutation test
We manually curated more than 5.5 million cells from 111 human tissues panels and 1,355 TCs from 61 adult and fetal tissues across 12 human organ systems (11 organ systems + sensory system). We used our t-statistic method to obtain the tissue-cell-type signatures panel by tissue. To construct a biological meaningful gene set for permutation shuffle, we curated ~5,600 genome-wide association studies (GWAS) and ~3,700 rare-variants association phenotypes from UK BioBank, GWAS catalog, and Genebass for trait-associated gene sets (TAGs). After stringent quality control criteria, we permutated the null model as a matrix of 19,663 TAGs by 1,355 TCs. For each inquiry gene list, we conduct CSEA across 1,355 TCs and get the permutated p-values by ranking among 19,663 TAGs and 1,355 TCs. Then we use Fisher’s method to combine an overall p-value from the permutated p-values by TAGs and by TCs.
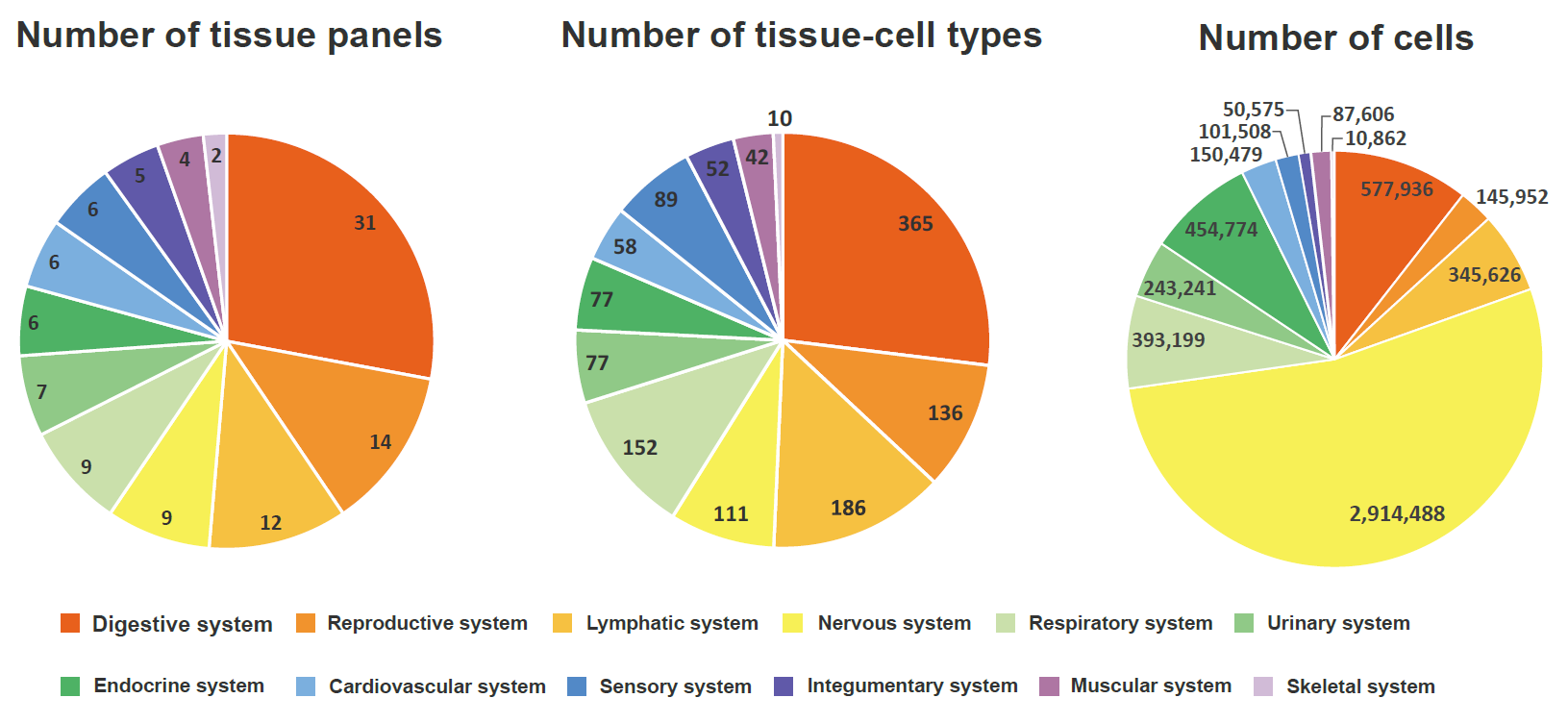
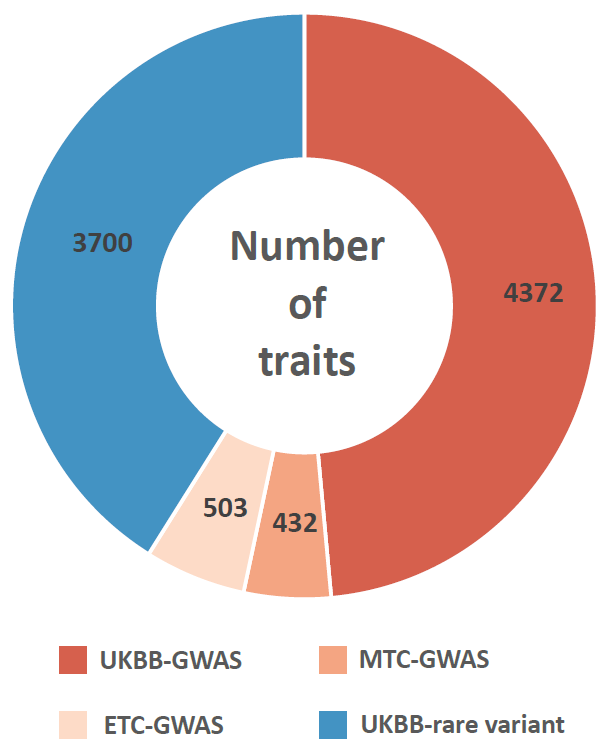
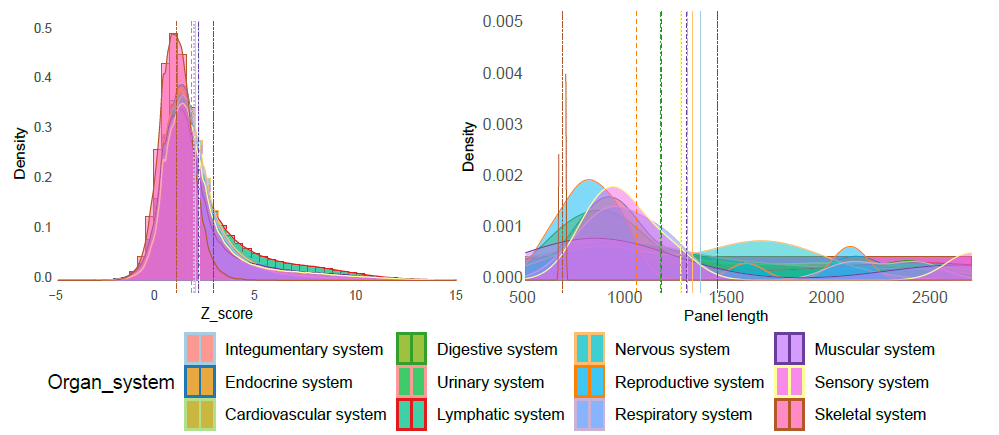
Sources
*WebCSEA is free and open to all users and there is no login requirement.



