Gene Page: CHAF1A
Summary ?
| GeneID | 10036 |
| Symbol | CHAF1A |
| Synonyms | CAF-1|CAF1|CAF1B|CAF1P150|P150 |
| Description | chromatin assembly factor 1 subunit A |
| Reference | MIM:601246|HGNC:HGNC:1910|Ensembl:ENSG00000167670|HPRD:03148|Vega:OTTHUMG00000181922 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.218 |
| Fetal beta | 0.763 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0064 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04346879 | 19 | 4402785 | CHAF1A | 1.325E-4 | -0.214 | 0.03 | DMG:Wockner_2014 |
| ch.19.239668R | 19 | 4434073 | CHAF1A | 2.035E-4 | -0.599 | 0.035 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12462529 | 19 | 4359525 | CHAF1A | ENSG00000167670.11 | 7.11E-7 | 0.01 | -43134 | gtex_brain_ba24 |
| rs7769 | 19 | 4360543 | CHAF1A | ENSG00000167670.11 | 9.539E-7 | 0.01 | -42116 | gtex_brain_ba24 |
| rs11670201 | 19 | 4364777 | CHAF1A | ENSG00000167670.11 | 1.026E-6 | 0.01 | -37882 | gtex_brain_ba24 |
| rs11666453 | 19 | 4368797 | CHAF1A | ENSG00000167670.11 | 9.384E-7 | 0.01 | -33862 | gtex_brain_ba24 |
| rs56033249 | 19 | 4369447 | CHAF1A | ENSG00000167670.11 | 9.347E-7 | 0.01 | -33212 | gtex_brain_ba24 |
| rs11667543 | 19 | 4374850 | CHAF1A | ENSG00000167670.11 | 8.805E-7 | 0.01 | -27809 | gtex_brain_ba24 |
| rs11085073 | 19 | 4375162 | CHAF1A | ENSG00000167670.11 | 9.446E-7 | 0.01 | -27497 | gtex_brain_ba24 |
| rs62129349 | 19 | 4385659 | CHAF1A | ENSG00000167670.11 | 4.372E-7 | 0.01 | -17000 | gtex_brain_ba24 |
| rs11085074 | 19 | 4386352 | CHAF1A | ENSG00000167670.11 | 4.368E-7 | 0.01 | -16307 | gtex_brain_ba24 |
| rs55637375 | 19 | 4386845 | CHAF1A | ENSG00000167670.11 | 4.366E-7 | 0.01 | -15814 | gtex_brain_ba24 |
| rs62129351 | 19 | 4387125 | CHAF1A | ENSG00000167670.11 | 6.666E-7 | 0.01 | -15534 | gtex_brain_ba24 |
| rs9973266 | 19 | 4392715 | CHAF1A | ENSG00000167670.11 | 5.636E-7 | 0.01 | -9944 | gtex_brain_ba24 |
| rs62129353 | 19 | 4395763 | CHAF1A | ENSG00000167670.11 | 4.619E-7 | 0.01 | -6896 | gtex_brain_ba24 |
| rs62129354 | 19 | 4396505 | CHAF1A | ENSG00000167670.11 | 4.441E-7 | 0.01 | -6154 | gtex_brain_ba24 |
| rs62129355 | 19 | 4397034 | CHAF1A | ENSG00000167670.11 | 4.687E-7 | 0.01 | -5625 | gtex_brain_ba24 |
| rs62129356 | 19 | 4400314 | CHAF1A | ENSG00000167670.11 | 3.34E-7 | 0.01 | -2345 | gtex_brain_ba24 |
| rs2230636 | 19 | 4409756 | CHAF1A | ENSG00000167670.11 | 1.114E-7 | 0.01 | 7097 | gtex_brain_ba24 |
| rs56411083 | 19 | 4410646 | CHAF1A | ENSG00000167670.11 | 1.062E-7 | 0.01 | 7987 | gtex_brain_ba24 |
| rs34080966 | 19 | 4417845 | CHAF1A | ENSG00000167670.11 | 8.978E-8 | 0.01 | 15186 | gtex_brain_ba24 |
| rs56227586 | 19 | 4418515 | CHAF1A | ENSG00000167670.11 | 9.793E-8 | 0.01 | 15856 | gtex_brain_ba24 |
| rs56272533 | 19 | 4420042 | CHAF1A | ENSG00000167670.11 | 1.004E-7 | 0.01 | 17383 | gtex_brain_ba24 |
| rs56088814 | 19 | 4422178 | CHAF1A | ENSG00000167670.11 | 1.585E-7 | 0.01 | 19519 | gtex_brain_ba24 |
| rs11668886 | 19 | 4423419 | CHAF1A | ENSG00000167670.11 | 5.083E-7 | 0.01 | 20760 | gtex_brain_ba24 |
| rs59488897 | 19 | 4425236 | CHAF1A | ENSG00000167670.11 | 8.978E-8 | 0.01 | 22577 | gtex_brain_ba24 |
| rs55660045 | 19 | 4425305 | CHAF1A | ENSG00000167670.11 | 8.978E-8 | 0.01 | 22646 | gtex_brain_ba24 |
| rs62130978 | 19 | 4429007 | CHAF1A | ENSG00000167670.11 | 9.793E-8 | 0.01 | 26348 | gtex_brain_ba24 |
| rs62130979 | 19 | 4432444 | CHAF1A | ENSG00000167670.11 | 8.858E-8 | 0.01 | 29785 | gtex_brain_ba24 |
| rs12459919 | 19 | 4434503 | CHAF1A | ENSG00000167670.11 | 8.978E-8 | 0.01 | 31844 | gtex_brain_ba24 |
| rs11666856 | 19 | 4437450 | CHAF1A | ENSG00000167670.11 | 3.259E-7 | 0.01 | 34791 | gtex_brain_ba24 |
| rs11669263 | 19 | 4441407 | CHAF1A | ENSG00000167670.11 | 8.978E-8 | 0.01 | 38748 | gtex_brain_ba24 |
| rs56156435 | 19 | 4444654 | CHAF1A | ENSG00000167670.11 | 3.568E-7 | 0.01 | 41995 | gtex_brain_ba24 |
| rs760369 | 19 | 4449287 | CHAF1A | ENSG00000167670.11 | 9.908E-8 | 0.01 | 46628 | gtex_brain_ba24 |
| rs1127888 | 19 | 4454083 | CHAF1A | ENSG00000167670.11 | 1.592E-7 | 0.01 | 51424 | gtex_brain_ba24 |
| rs72990643 | 19 | 4454736 | CHAF1A | ENSG00000167670.11 | 1.586E-7 | 0.01 | 52077 | gtex_brain_ba24 |
| rs12459922 | 19 | 4455862 | CHAF1A | ENSG00000167670.11 | 1.59E-7 | 0.01 | 53203 | gtex_brain_ba24 |
| rs56153949 | 19 | 4457518 | CHAF1A | ENSG00000167670.11 | 4.703E-8 | 0.01 | 54859 | gtex_brain_ba24 |
| rs11670503 | 19 | 4458063 | CHAF1A | ENSG00000167670.11 | 2.198E-7 | 0.01 | 55404 | gtex_brain_ba24 |
| rs12462529 | 19 | 4359525 | CHAF1A | ENSG00000167670.11 | 1.93E-7 | 0 | -43134 | gtex_brain_putamen_basal |
| rs7769 | 19 | 4360543 | CHAF1A | ENSG00000167670.11 | 3.706E-8 | 0 | -42116 | gtex_brain_putamen_basal |
| rs62129346 | 19 | 4363289 | CHAF1A | ENSG00000167670.11 | 3.21E-10 | 0 | -39370 | gtex_brain_putamen_basal |
| rs11670201 | 19 | 4364777 | CHAF1A | ENSG00000167670.11 | 3.732E-8 | 0 | -37882 | gtex_brain_putamen_basal |
| rs732716 | 19 | 4366219 | CHAF1A | ENSG00000167670.11 | 1.295E-9 | 0 | -36440 | gtex_brain_putamen_basal |
| rs11085072 | 19 | 4368142 | CHAF1A | ENSG00000167670.11 | 2.201E-7 | 0 | -34517 | gtex_brain_putamen_basal |
| rs11666453 | 19 | 4368797 | CHAF1A | ENSG00000167670.11 | 3.634E-8 | 0 | -33862 | gtex_brain_putamen_basal |
| rs56033249 | 19 | 4369447 | CHAF1A | ENSG00000167670.11 | 3.571E-8 | 0 | -33212 | gtex_brain_putamen_basal |
| rs11667543 | 19 | 4374850 | CHAF1A | ENSG00000167670.11 | 3.063E-8 | 0 | -27809 | gtex_brain_putamen_basal |
| rs11085073 | 19 | 4375162 | CHAF1A | ENSG00000167670.11 | 3.026E-8 | 0 | -27497 | gtex_brain_putamen_basal |
| rs62129349 | 19 | 4385659 | CHAF1A | ENSG00000167670.11 | 8.867E-11 | 0 | -17000 | gtex_brain_putamen_basal |
| rs11085074 | 19 | 4386352 | CHAF1A | ENSG00000167670.11 | 8.697E-11 | 0 | -16307 | gtex_brain_putamen_basal |
| rs55637375 | 19 | 4386845 | CHAF1A | ENSG00000167670.11 | 8.576E-11 | 0 | -15814 | gtex_brain_putamen_basal |
| rs62129351 | 19 | 4387125 | CHAF1A | ENSG00000167670.11 | 1.226E-8 | 0 | -15534 | gtex_brain_putamen_basal |
| rs9973266 | 19 | 4392715 | CHAF1A | ENSG00000167670.11 | 1.16E-8 | 0 | -9944 | gtex_brain_putamen_basal |
| rs5826842 | 19 | 4393294 | CHAF1A | ENSG00000167670.11 | 2.983E-10 | 0 | -9365 | gtex_brain_putamen_basal |
| rs62129353 | 19 | 4395763 | CHAF1A | ENSG00000167670.11 | 7.121E-9 | 0 | -6896 | gtex_brain_putamen_basal |
| rs62129354 | 19 | 4396505 | CHAF1A | ENSG00000167670.11 | 6.494E-9 | 0 | -6154 | gtex_brain_putamen_basal |
| rs62129355 | 19 | 4397034 | CHAF1A | ENSG00000167670.11 | 4.58E-11 | 0 | -5625 | gtex_brain_putamen_basal |
| rs62129356 | 19 | 4400314 | CHAF1A | ENSG00000167670.11 | 1.838E-9 | 0 | -2345 | gtex_brain_putamen_basal |
| rs200677191 | 19 | 4402659 | CHAF1A | ENSG00000167670.11 | 7.349E-10 | 0 | 0 | gtex_brain_putamen_basal |
| rs2230636 | 19 | 4409756 | CHAF1A | ENSG00000167670.11 | 1.197E-9 | 0 | 7097 | gtex_brain_putamen_basal |
| rs56411083 | 19 | 4410646 | CHAF1A | ENSG00000167670.11 | 6.103E-10 | 0 | 7987 | gtex_brain_putamen_basal |
| rs34080966 | 19 | 4417845 | CHAF1A | ENSG00000167670.11 | 1.034E-9 | 0 | 15186 | gtex_brain_putamen_basal |
| rs56227586 | 19 | 4418515 | CHAF1A | ENSG00000167670.11 | 5.537E-10 | 0 | 15856 | gtex_brain_putamen_basal |
| rs56272533 | 19 | 4420042 | CHAF1A | ENSG00000167670.11 | 1.135E-9 | 0 | 17383 | gtex_brain_putamen_basal |
| rs56088814 | 19 | 4422178 | CHAF1A | ENSG00000167670.11 | 6.092E-10 | 0 | 19519 | gtex_brain_putamen_basal |
| rs11668886 | 19 | 4423419 | CHAF1A | ENSG00000167670.11 | 9.97E-10 | 0 | 20760 | gtex_brain_putamen_basal |
| rs59488897 | 19 | 4425236 | CHAF1A | ENSG00000167670.11 | 1.034E-9 | 0 | 22577 | gtex_brain_putamen_basal |
| rs55660045 | 19 | 4425305 | CHAF1A | ENSG00000167670.11 | 1.034E-9 | 0 | 22646 | gtex_brain_putamen_basal |
| rs62130978 | 19 | 4429007 | CHAF1A | ENSG00000167670.11 | 1.03E-9 | 0 | 26348 | gtex_brain_putamen_basal |
| rs62130979 | 19 | 4432444 | CHAF1A | ENSG00000167670.11 | 5.601E-10 | 0 | 29785 | gtex_brain_putamen_basal |
| rs12459919 | 19 | 4434503 | CHAF1A | ENSG00000167670.11 | 1.034E-9 | 0 | 31844 | gtex_brain_putamen_basal |
| rs11666856 | 19 | 4437450 | CHAF1A | ENSG00000167670.11 | 1.422E-11 | 0 | 34791 | gtex_brain_putamen_basal |
| rs11669263 | 19 | 4441407 | CHAF1A | ENSG00000167670.11 | 1.034E-9 | 0 | 38748 | gtex_brain_putamen_basal |
| rs56156435 | 19 | 4444654 | CHAF1A | ENSG00000167670.11 | 2.381E-11 | 0 | 41995 | gtex_brain_putamen_basal |
| rs78869060 | 19 | 4444742 | CHAF1A | ENSG00000167670.11 | 5.083E-11 | 0 | 42083 | gtex_brain_putamen_basal |
| rs760369 | 19 | 4449287 | CHAF1A | ENSG00000167670.11 | 7.253E-12 | 0 | 46628 | gtex_brain_putamen_basal |
| rs1127888 | 19 | 4454083 | CHAF1A | ENSG00000167670.11 | 1.417E-11 | 0 | 51424 | gtex_brain_putamen_basal |
| rs72990643 | 19 | 4454736 | CHAF1A | ENSG00000167670.11 | 1.418E-11 | 0 | 52077 | gtex_brain_putamen_basal |
| rs12459922 | 19 | 4455862 | CHAF1A | ENSG00000167670.11 | 1.414E-11 | 0 | 53203 | gtex_brain_putamen_basal |
| rs56153949 | 19 | 4457518 | CHAF1A | ENSG00000167670.11 | 1.134E-9 | 0 | 54859 | gtex_brain_putamen_basal |
| rs11670503 | 19 | 4458063 | CHAF1A | ENSG00000167670.11 | 1.475E-11 | 0 | 55404 | gtex_brain_putamen_basal |
| rs143191932 | 19 | 4459785 | CHAF1A | ENSG00000167670.11 | 6.021E-7 | 0 | 57126 | gtex_brain_putamen_basal |
| rs149060351 | 19 | 4460005 | CHAF1A | ENSG00000167670.11 | 7.108E-7 | 0 | 57346 | gtex_brain_putamen_basal |
| rs11669260 | 19 | 4460272 | CHAF1A | ENSG00000167670.11 | 8.009E-7 | 0 | 57613 | gtex_brain_putamen_basal |
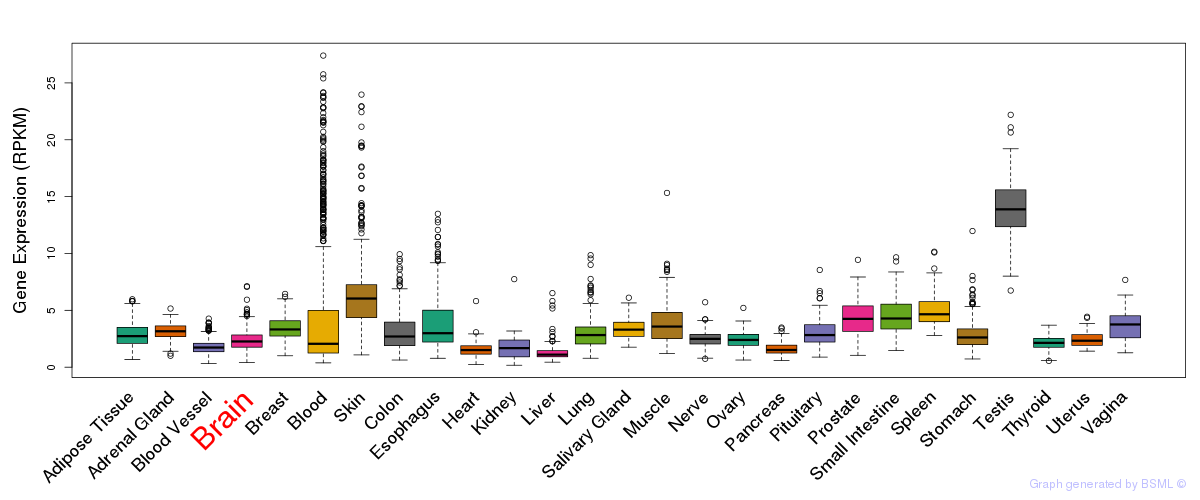
Section II. Transcriptome annotation
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003682 | chromatin binding | TAS | 7600578 | |
| GO:0051082 | unfolded protein binding | TAS | 7600578 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006461 | protein complex assembly | TAS | 7600578 | |
| GO:0006335 | DNA replication-dependent nucleosome assembly | TAS | 7600578 | |
| GO:0006281 | DNA repair | IEA | - | |
| GO:0006260 | DNA replication | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005678 | chromatin assembly complex | TAS | 7600578 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASF1A | CGI-98 | CIA | DKFZp547E2110 | HSPC146 | ASF1 anti-silencing function 1 homolog A (S. cerevisiae) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11897662 |
| ASF1B | CIA-II | FLJ10604 | ASF1 anti-silencing function 1 homolog B (S. cerevisiae) | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 11897662 |17353931 |
| BAZ1B | WBSCR10 | WBSCR9 | WSTF | bromodomain adjacent to zinc finger domain, 1B | - | HPRD | 12837248 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | Affinity Capture-Western Far Western Two-hybrid | BioGRID | 15143166 |
| CBX1 | CBX | HP1-BETA | HP1Hs-beta | HP1Hsbeta | M31 | MOD1 | p25beta | chromobox homolog 1 (HP1 beta homolog Drosophila ) | - | HPRD | 10549285 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | - | HPRD | 10549285 |10938122 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10938122 |12697822 |
| CHAF1B | CAF-1 | CAF-IP60 | CAF1 | CAF1A | CAF1P60 | MPHOSPH7 | MPP7 | chromatin assembly factor 1, subunit B (p60) | - | HPRD,BioGRID | 9614144 |
| HIST1H3E | H3.1 | H3/d | H3FD | histone cluster 1, H3e | CAF-1 p150 interacts with H3.1. This interaction was modeled on a demonstrated interaction between human CAF-1 p150 and H3.1 from an unspecified species. | BIND | 15664198 |
| MBD1 | CXXC3 | PCM1 | RFT | methyl-CpG binding domain protein 1 | - | HPRD,BioGRID | 12697822 |
| MBD1 | CXXC3 | PCM1 | RFT | methyl-CpG binding domain protein 1 | MBD1 interacts with CAF-1. | BIND | 15327775 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD,BioGRID | 10648606 |
| SETDB1 | ESET | KG1T | KIAA0067 | KMT1E | SET domain, bifurcated 1 | SETDB1 interacts with CAF-1. | BIND | 15327775 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 15456888 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS DN | 139 | 76 | All SZGR 2.0 genes in this pathway |
| ODONNELL TARGETS OF MYC AND TFRC DN | 45 | 25 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN MELANOMA RELAPSE UP | 61 | 25 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA SUBGROUPS | 30 | 20 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY UP | 70 | 47 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND MACROPHAGE | 77 | 50 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN | 98 | 69 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G1 S | 147 | 76 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |