 |
||||||
|
|
||||||
Data Source for Linkage Study |
| 1. Method |
|---|
|
The linkage datasets are collected based
on two genome scan meta-analyses (GSMA). The first GSMA was
applied to data from 20 schizophrenia genome-wide linkage scans (
Lewis et al. 2003). We selected 12 bins whose
PAvgRnk
and Pord
are both <0.05, identified their corresponding physical locations
on chromosomes, and extracted genes within these regions. These
genes are then defined as " GSMA_I"
in our database. The second GSMA was applied to 32 schizophrenia
genome scans ( Ng et al. 2009). We obtained genes from 10
bins whose PSR
are <0.05 for all the samples and 6 bins only for European
samples. These two lists are defined as " GSMA_IIA"
and " GSMA_IIE".
The P value of each linkage bin was assigned to the genes
within the bin. |
| 2. Dataset Description |
| Table 1. Composition of linkage study defined gene set. |
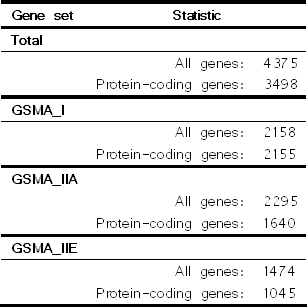
|
| Table 2. Overlap between linkage gene sets. |

|
| References |
|
| Copyright © Bioinformatics and Systems Medicine Laboratory All Rights Reserved since 2009. |