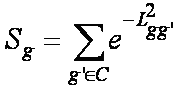|
||||||
|
|
||||||
Data Source for Gene Network Dataset |
| 1. Data collection |
|---|
|
Gene networks play important role in causing complex diseases. To examine schizophrenia candidate genes in the context of protein-protein interaction network, we first constructed a comprehensive human PPI network by integrating protein-protein interaction pairs from six databases: Human Protein Reference Database (HPRD) ( Peri et al. 2003), BIND ( Bader et al. 2001), IntAct ( Hermjakob et al. 2004), MINT ( Zanzoni et al. 2002), Reactome ( Matthews et al. 2009) and DIP ( Salwinski et al. 2004). We then map our core genes onto this PPI network and found 32 of the core genes are present at the network. In network topology, proteins in the shortest path tend to have same or similar biological process (Managbanag et al. 2008). Thus we identified the shortest paths between any two of the 32 core genes. Finally, we included all the genes whose coding proteins are present at these shortest paths. A total of 1035 genes were selected based on this network feature. |
| 2. Scoring system |
|
Each gene in the shortest path to a pair of core genes was assigned a score to describe it closeness to the phenotype (schizophrenia in this case). We modified Wu et al. (2008) method to calculate the closeness of a gene in the shortest path to schizophrenia (i.e. core genes). The closeness of a gene g in the shortest path to a schizophrenia core gene is calculated by Gaussian kernel where g' is the core gene and L is the distance between genes g and g' in the shortest path. Therefore, the final score of the gene is the sum of its closeness to all core genes:
where C is the set of core genes. |
| 3. Score distribution |
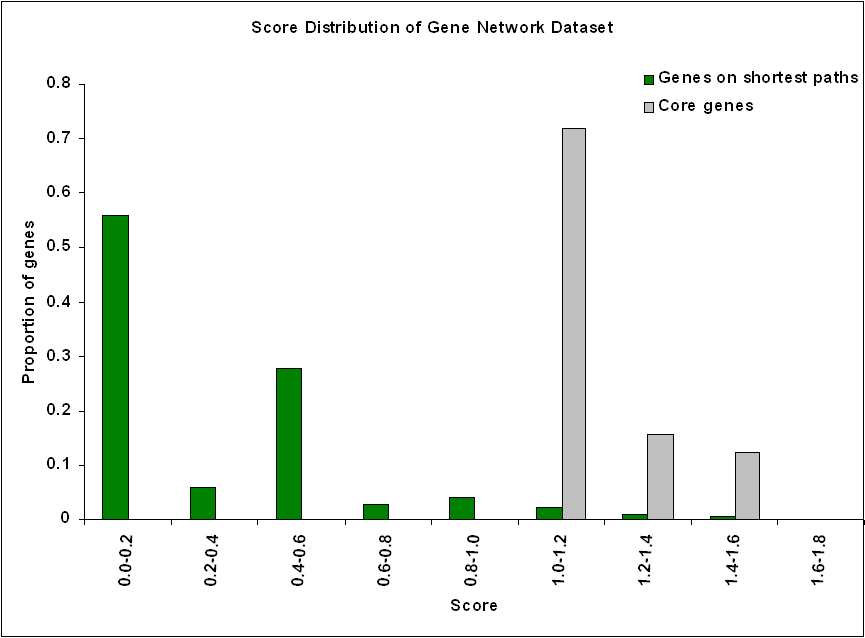 |
| Figure 1. Score distribution of PPI network dataset |
| References |
|
| Copyright © Bioinformatics and Systems Medicine Laboratory All Rights Reserved since 2009. |