Genome-wide association studies
| Study ID | # SNPs | # Genes | Tier | Reference | Inside | Update |
|---|---|---|---|---|---|---|
| CV:GWAScat | 560 | 486 | Tier 1 | NHGRI-EBI GWAS Catalog | SZGR 2.0 page | 05/18/2016 |
| CV:GWASdb | 560 | 2130 | Tier 1 | GWASdb | SZGR 2.0 page | 05/04/2016 |
| CV:PGC128 | 128 | 148 | Tier 1 | Psychiatric Genomics Consortium (PGC) | SZGR 2.0 page | 06/01/2016 |
| CV:PheWAS | 128 | 148 | Tier 1 | PheWAS | SZGR 2.0 page | 06/01/2016 |
| CV:Ripke_2013 | 128 | 27 | Tier 1 | PubMed | 06/01/2016 | |
| CV:PGCnp | 128 | 27 | Tier 2 | Psychiatric Genomics Consortium (PGC) | PubMed | 06/01/2016 |
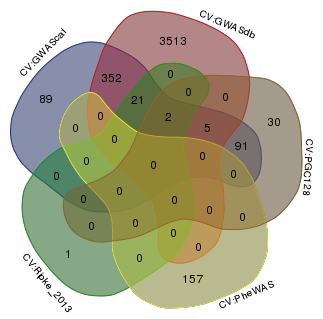
Venn diagram comparing the SNPs among the five Tier 1 SNPs.
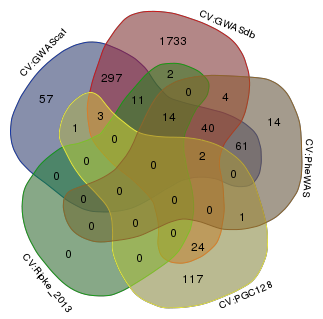
Venn diagram comparing the genes mapped to Tier 1 SNPs within 50kb.
De novo mutations
| Study ID | First author | Year | Journal | Sample size | Platform | Sequencer | PMID |
|---|---|---|---|---|---|---|---|
| DNM:Guipponi_2014 | Guipponi | 2014 | PLoS One | 53 | Agilent SureSelect Human ALL Exon kits | HiSeq | 25420024 |
| DNM:Girard_2011 | Girard | 2011 | Nature Genetics | 623 | Agilent SureSelect Human ALL Exon kits | HiSeq | 21743468 |
| DNM:Takata_2014 | Takata A | 2014 | Neuron | 231 case and 34 control trios | Agilent SureSelect v2 (n = 85 trios), NimbleGen SeqCap EZ v2 (n = 180 trios) | HiSeq | 24853937 |
| DNM:McCarthy_2014 | McCarthy | 2014 | Molecular Psychiatry | 57 | NimbleGen SeqCap EZ Human Exome Library v2.0 probes | HiSeq (101 bp PE reads) | 24776741 |
| DNM:Fromer_2014 | Fromer | 2014 | Nature | 617 | Agilent SureSelect Human All Exon v.2, Nimblegen SeqCap EZ Human Exome Library v2.0, Agilent SureSelect Human All Exon 50MB | HiSeq (76 bp, 101 bp, 76 bp PE reads) | 24463507 |
| DNM:Gulsuner_2013 | Gulsuner | 2013 | Cell | 105 | NimbleGen SeqCap EZ Human Exome Library v2.0 | HiSeq (101 bp PE reads) | 23911319 |
| DNM:Xu_2012 | Xu | 2012 | Nature Genetics | 231 | Agilent SureSelect v2 (n = 85 trios), NimbleGen SeqCap EZ v2 (n = 180 trios) | HiSeq | 23042115 |
| DNM:Xu_2011 | Xu | 2011 | Nature Genetics | 53 | Agilent SureSelect Human All Exon Target Enrichment System | HiSeq (50 bp PE reads) | 21822266 |
| DNM:Awadalla_2010 | Awadalla | 2010 | AJHG | ASD and SCZ cases (n = 285) and 285 controls | Targeted sequencing | 401 synapse-expressed genes | 20797689 |
| Footnote: The data in DNM:Xu_2011 has been included in DNM:Xu_2012. | |||||||
Differentially expressed genes
| Study ID | Platform | Description | # DEGs | Tissue | In SZGR2 | Reference |
|---|---|---|---|---|---|---|
| DEG:Zhao_2015 | RNA-sequencing | 35 schizophrenia and 35 controls | 105 genes | Brain | Included | 25113377 |
| DEG:Sanders_2014 | Illumina HT12v4 | 413 schizophrenia and 446 controls | 95 transcripts | Lymphoblastoid cell lines | Included | 23904455 |
Differentially methylated genes
| Study ID | Platform | # CpGs | Description | # DMGs | Tissue | In SZGR2 | Reference |
|---|---|---|---|---|---|---|---|
| DMG:Montano_2016 | Illumina HumanMethylation450 BeadChip | 456,513 CpG | 689 schizophrenia and 645 controls | 172 records | Included | 27074206 | |
| DMG:Wockner_2014 | Illumina HumanMethylation450 BeadChip | 485,000 CpG | 24 schizophrenia and 24 controls | 4641 | Blood | Included | 26809779 |
| DMG:Nishioka_2013 | Illumina Infinium HumanMethylation27 BeadChip | 27,578 CpG | 18 Japanese FESZ patients (11 males and 7 females); 15 Japanese age-matched healthy controls (10 males and 5 females) | 603 | Blood | Included | 23235336 |
| DMG:Jaffe_2016 | Illumina HumanMethylation450 BeadChip | 485,000 CpG | 108 schizophrenia and 136 controls | 2104 | DLPFC | Included | 26619358 |
| DMG:vanEijk_2014 | Illumina Infinium 27k Human DNA methylation Beadchip v1.2 | 27,000 CpGs | 260 SZ patients and 250 unaffected controls | Blood | Included | 25424713 | |
Copy number variation (CNV) loci in schizophrenia
| Locus | Studies | References | Replication | # genes | ||||
|---|---|---|---|---|---|---|---|---|
| 1q21.1 del | CNV:Levinson_2011 | 21285140 | Yes | 35 | ||||
| CNV:Rees_2014 | 24311552 | |||||||
| 2p16.3 | CNV:Levinson_2011 | 21285140 | Yes | 1 | ||||
| CNV:Rees_2014 | 24311552 | |||||||
| 3q29 | CNV:Levinson_2011 | 21285140 | Yes | 21 | ||||
| CNV:Rees_2014 | 24311552 | |||||||
| 7q11.23 | CNV:Sanders_2011 | 21658581 | Yes | 26 | ||||
| CNV:Rees_2014 | 24311552 | |||||||
| 7q36.3 | CNV:Levinson_2011 | 21285140 | Yes | 1 | ||||
| CNV:Rees_2014 | 24311552 | |||||||
| 15q11.2 | CNV:Rees_2014 | 24311552 | No | 14 | ||||
| AS/PWS dup | CNV:Rees_2014 | 24311552 | No | 1 | ||||
| 15q13.3 del | CNV:Levinson_2011 | 21285140 | Yes | 16 | ||||
| CNV:Rees_2014 | 24311552 | |||||||
| 16p13.11 dup | CNV:Rees_2014 | 21285140 | No | 9 | ||||
| 16p11.2 dup | CNV:Levinson_2011 | 21285140 | Yes | 27 | ||||
| CNV:Rees_2014 | 24311552 | |||||||
| 17q12 del | CNV:Levinson_2011 | 21285140 | Yes | 17 | ||||
| CNV:Rees_2014 | 24311552 | |||||||
| 22q11.21 del | CNV:Levinson_2011 | 21285140 | Yes | 49 | ||||
| CNV:Rees_2014 | 24311552 | |||||||