Gene Page: EDDM3A
Summary ?
| GeneID | 10876 |
| Symbol | EDDM3A |
| Synonyms | EP3A|FAM12A|HE3-ALPHA|HE3A|HE3ALPHA|RAM1 |
| Description | epididymal protein 3A |
| Reference | MIM:611580|HGNC:HGNC:16978|Ensembl:ENSG00000181562|HPRD:13289|Vega:OTTHUMG00000029581 |
| Gene type | protein-coding |
| Map location | 14q11.2 |
| Pascal p-value | 0.394 |
| Fetal beta | -0.16 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
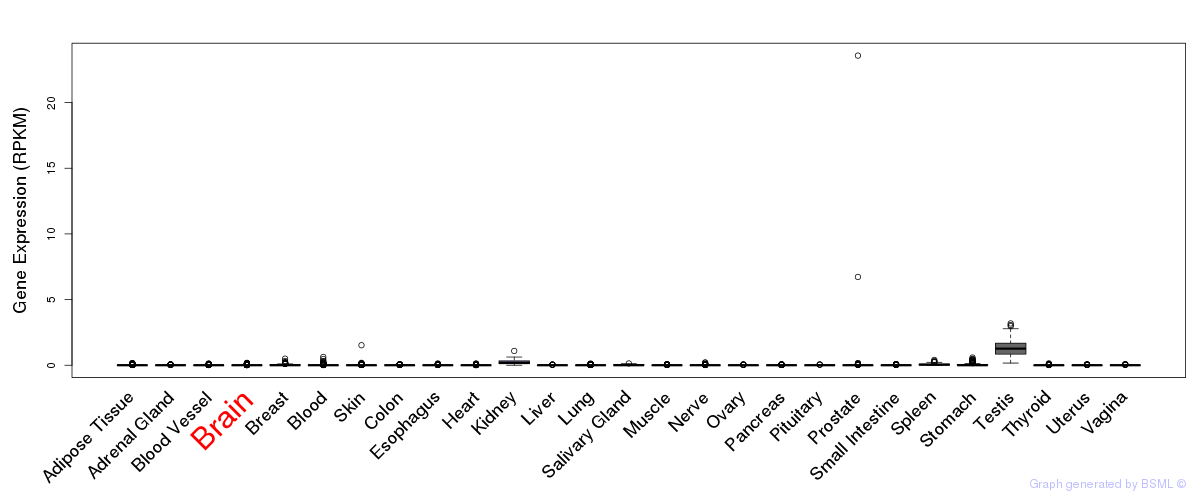
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACTR10 | 0.86 | 0.87 |
| C10orf97 | 0.85 | 0.86 |
| VAPA | 0.84 | 0.87 |
| MRPS35 | 0.84 | 0.84 |
| ORC4L | 0.84 | 0.84 |
| RAP1GDS1 | 0.83 | 0.87 |
| MORF4L1 | 0.82 | 0.84 |
| CCNC | 0.82 | 0.82 |
| ATG5 | 0.82 | 0.83 |
| POLR3F | 0.82 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.58 | -0.53 |
| AF347015.8 | -0.56 | -0.50 |
| AF347015.26 | -0.55 | -0.50 |
| GPT | -0.55 | -0.56 |
| MT-CO2 | -0.55 | -0.49 |
| MT-CYB | -0.54 | -0.49 |
| AF347015.15 | -0.54 | -0.49 |
| AF347015.18 | -0.54 | -0.53 |
| AF347015.33 | -0.53 | -0.49 |
| EIF4EBP3 | -0.53 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0004522 | pancreatic ribonuclease activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007321 | sperm displacement | TAS | 7514008 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | TAS | 7514008 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |