Gene Page: CTNNB1
Summary ?
| GeneID | 1499 |
| Symbol | CTNNB1 |
| Synonyms | CTNNB|MRD19|armadillo |
| Description | catenin beta 1 |
| Reference | MIM:116806|HGNC:HGNC:2514|Ensembl:ENSG00000168036|HPRD:00286|Vega:OTTHUMG00000131393 |
| Gene type | protein-coding |
| Map location | 3p21 |
| Pascal p-value | 0.636 |
| Fetal beta | -0.341 |
| DMG | 1 (# studies) |
| eGene | Putamen basal ganglia |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.9379 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04180460 | 3 | 41240962 | CTNNB1 | 4.47E-10 | -0.013 | 8.02E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs55840513 | 3 | 41239045 | CTNNB1 | ENSG00000168036.12 | 6.36481E-6 | 0.05 | 2717 | gtex_brain_putamen_basal |
| rs13075993 | 3 | 41260017 | CTNNB1 | ENSG00000168036.12 | 6.68607E-6 | 0.05 | 23689 | gtex_brain_putamen_basal |
| rs3774369 | 3 | 41267525 | CTNNB1 | ENSG00000168036.12 | 6.73806E-6 | 0.05 | 31197 | gtex_brain_putamen_basal |
| rs11564454 | 3 | 41274477 | CTNNB1 | ENSG00000168036.12 | 6.64323E-6 | 0.05 | 38149 | gtex_brain_putamen_basal |
| rs3774371 | 3 | 41276166 | CTNNB1 | ENSG00000168036.12 | 6.67766E-6 | 0.05 | 39838 | gtex_brain_putamen_basal |
| rs11564465 | 3 | 41277040 | CTNNB1 | ENSG00000168036.12 | 6.67766E-6 | 0.05 | 40712 | gtex_brain_putamen_basal |
| rs150512515 | 3 | 41281409 | CTNNB1 | ENSG00000168036.12 | 6.67626E-6 | 0.05 | 45081 | gtex_brain_putamen_basal |
| rs11129895 | 3 | 41283382 | CTNNB1 | ENSG00000168036.12 | 6.68625E-6 | 0.05 | 47054 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
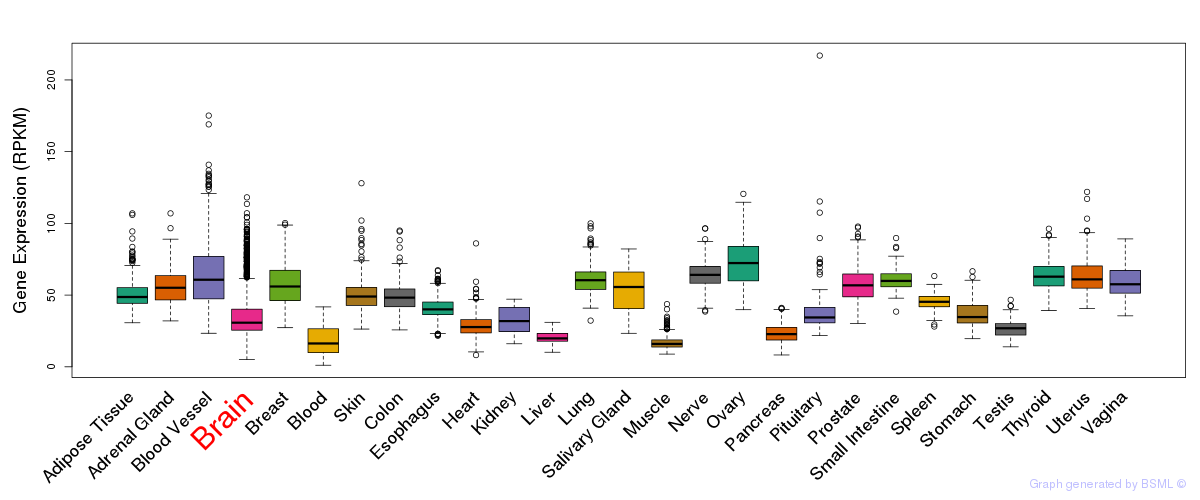
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | NAS | 10192393 | |
| GO:0003713 | transcription coactivator activity | NAS | 15572661 | |
| GO:0005198 | structural molecule activity | IEA | - | |
| GO:0008022 | protein C-terminus binding | IPI | 10773885 | |
| GO:0050681 | androgen receptor binding | NAS | 15572661 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001837 | epithelial to mesenchymal transition | TAS | 14679171 | |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0030521 | androgen receptor signaling pathway | NAS | 15572661 | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | NAS | 15572661 | |
| GO:0060070 | Wnt receptor signaling pathway through beta-catenin | IC | 9601641 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 11955436 |12000790 |12820959 |15327769 |16753179 | |
| GO:0005856 | cytoskeleton | TAS | 7806582 | |
| GO:0005634 | nucleus | TAS | 9065401 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 10837025 | |
| GO:0016328 | lateral plasma membrane | IDA | 12072559 | |
| GO:0030877 | beta-catenin destruction complex | IDA | 9601641 |16188939 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACP1 | HAAP | MGC111030 | MGC3499 | acid phosphatase 1, soluble | - | HPRD,BioGRID | 12438242 |
| AJAP1 | MOT8 | RP3-426F10.1 | SHREW-1 | SHREW1 | adherens junctions associated protein 1 | - | HPRD,BioGRID | 14595118 |
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | APC interacts with Beta-catenin. | BIND | 15525529 |
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | - | HPRD | 12000790 |
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | Beta-catenin interacts with APC. This interaction was modelled on a demonstrated interaction between human beta-catenin and mouse Apc. | BIND | 15294866 |
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | Affinity Capture-Western Co-crystal Structure Far Western Reconstituted Complex | BioGRID | 8259519 |9286858 |11251183 |11533658 |11707392 |11712088 |11972058 |12628243 |
| APC2 | APCL | adenomatosis polyposis coli 2 | - | HPRD,BioGRID | 9823329 |10021369 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 11916967 |
| ARHGEF4 | ASEF | ASEF1 | GEF4 | STM6 | Rho guanine nucleotide exchange factor (GEF) 4 | - | HPRD | 10947987 |
| AXIN1 | AXIN | MGC52315 | axin 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9734785 |12805222 |
| AXIN1 | AXIN | MGC52315 | axin 1 | - | HPRD | 12000790 |
| AXIN2 | AXIL | DKFZp781B0869 | MGC10366 | MGC126582 | axin 2 | - | HPRD,BioGRID | 10966653 |
| AXIN2 | AXIL | DKFZp781B0869 | MGC10366 | MGC126582 | axin 2 | beta-catenin interacts with the Axin2 promoter. | BIND | 15304487 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | CTNNB1 (beta-catenin) interacts with BCL3. | BIND | 15829968 |
| BCL9 | LGS | MGC131591 | B-cell CLL/lymphoma 9 | Beta-catenin interacts with LGS1. | BIND | 15294866 |
| BOC | - | Boc homolog (mouse) | - | HPRD,BioGRID | 12634428 |
| BTRC | BETA-TRCP | FBW1A | FBXW1 | FBXW1A | FWD1 | MGC4643 | bTrCP | bTrCP1 | betaTrCP | beta-transducin repeat containing | - | HPRD | 11818547 |12114015 |
| BTRC | BETA-TRCP | FBW1A | FBXW1 | FBXW1A | FWD1 | MGC4643 | bTrCP | bTrCP1 | betaTrCP | beta-transducin repeat containing | BTRC (Beta-TrCP1) interacts with CTNNB1 (Beta-catenin). | BIND | 15735746 |
| CA9 | CAIX | MN | carbonic anhydrase IX | - | HPRD,BioGRID | 14567991 |
| CACYBP | GIG5 | MGC87971 | PNAS-107 | RP1-102G20.6 | S100A6BP | SIP | calcyclin binding protein | Affinity Capture-Western | BioGRID | 11389839 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | - | HPRD | 9348292 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD | 12189238 |
| CBY1 | C22orf2 | CBY | HS508I15A | PGEA1 | PIGEA-14 | PIGEA14 | arb1 | chibby homolog 1 (Drosophila) | - | HPRD,BioGRID | 12712206 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | CTNNB1 (beta-catenin) interacts with the CCND1 promoter. | BIND | 15592430 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD | 11836555 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | CTNNB1 (beta-catenin) interacts with the CD82 (KAI1) promoter. | BIND | 15829968 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Affinity Capture-Western Co-crystal Structure | BioGRID | 7542250 |7876318 |7954478 |8227214 |9233779 |9405455 |9535896 |9819408 |10381631 |10772923 |11113628 |11245482 |11254878 |11348595 |11712088 |11960376 |12061792 |12169098 |12634428 |12640114 |12830000 |15023525 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | - | HPRD | 9512503 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Beta-catenin interacts with E-cadherin. This interaction was modelled on a demonstrated interaction between human beta-catenin and mouse E-cadherin. | BIND | 15294866 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | E-cadherin interacts with beta-catenin. | BIND | 14706341 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Beta-catenin interacts with E-cadherin. This interaction was modelled on a demonstrated interaction between beta-catenin from an unspecified species and human E-cadherin. | BIND | 15542433 |
| CDH10 | - | cadherin 10, type 2 (T2-cadherin) | Affinity Capture-Western | BioGRID | 10861224 |
| CDH11 | CAD11 | CDHOB | OB | OSF-4 | cadherin 11, type 2, OB-cadherin (osteoblast) | - | HPRD,BioGRID | 10029089 |
| CDH15 | CDH14 | CDH3 | CDHM | MCAD | cadherin 15, type 1, M-cadherin (myotubule) | - | HPRD | 9545347 |
| CDH16 | - | cadherin 16, KSP-cadherin | - | HPRD | 15023525 |
| CDH17 | CDH16 | FLJ26931 | HPT-1 | HPT1 | MGC138218 | MGC142024 | cadherin 17, LI cadherin (liver-intestine) | Affinity Capture-Western | BioGRID | 15023525 |
| CDH18 | CDH14 | CDH14L | CDH24 | EY-CADHERIN | cadherin 18, type 2 | Affinity Capture-Western | BioGRID | 10207020 |
| CDH2 | CD325 | CDHN | CDw325 | NCAD | cadherin 2, type 1, N-cadherin (neuronal) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12604612 |14625392 |
| CDH2 | CD325 | CDHN | CDw325 | NCAD | cadherin 2, type 1, N-cadherin (neuronal) | - | HPRD | 12151522 |
| CDH24 | CDH11L | FLJ25193 | MGC131880 | cadherin-like 24 | - | HPRD | 12734196 |
| CDH3 | CDHP | HJMD | PCAD | cadherin 3, type 1, P-cadherin (placental) | - | HPRD | 8227214 |10381631 |11889072 |12800191 |
| CDH3 | CDHP | HJMD | PCAD | cadherin 3, type 1, P-cadherin (placental) | Affinity Capture-Western Co-purification | BioGRID | 10381631 |10910767 |
| CDH5 | 7B4 | CD144 | FLJ17376 | cadherin 5, type 2 (vascular endothelium) | - | HPRD | 12413882 |
| CDH5 | 7B4 | CD144 | FLJ17376 | cadherin 5, type 2 (vascular endothelium) | Affinity Capture-Western | BioGRID | 9434630 |12003790 |
| CDH6 | KCAD | cadherin 6, type 2, K-cadherin (fetal kidney) | Affinity Capture-Western | BioGRID | 10207020 |
| CDH7 | CDH7L1 | cadherin 7, type 2 | - | HPRD,BioGRID | 10861224 |
| CDH8 | Nbla04261 | cadherin 8, type 2 | - | HPRD,BioGRID | 9521872 |
| CDH9 | MGC125386 | cadherin 9, type 2 (T1-cadherin) | - | HPRD,BioGRID | 10861224 |
| CDK5 | PSSALRE | cyclin-dependent kinase 5 | - | HPRD | 11168528 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11168528 |
| CDON | CDO | MGC111524 | ORCAM | Cdon homolog (mouse) | - | HPRD,BioGRID | 12634428 |
| CEBPA | C/EBP-alpha | CEBP | CCAAT/enhancer binding protein (C/EBP), alpha | - | HPRD,BioGRID | 12861022 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 11527961 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Phenotypic Enhancement | BioGRID | 15782138 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD | 12432063 |
| CTNNA1 | CAP102 | FLJ36832 | catenin (cadherin-associated protein), alpha 1, 102kDa | - | HPRD,BioGRID | 7706414 |
| CTNNA1 | CAP102 | FLJ36832 | catenin (cadherin-associated protein), alpha 1, 102kDa | Beta-catenin interacts with alpha-catenin. This interaction was modelled on a demonstrated interaction between human beta-catenin and mouse alpha-catenin. | BIND | 15294866 |
| CTNNA3 | MGC26194 | MGC75041 | VR22 | catenin (cadherin-associated protein), alpha 3 | - | HPRD,BioGRID | 11590244 |
| CTNNBIP1 | ICAT | MGC15093 | catenin, beta interacting protein 1 | - | HPRD,BioGRID | 10898789 |
| CTNND1 | CAS | CTNND | KIAA0384 | P120CAS | P120CTN | p120 | catenin (cadherin-associated protein), delta 1 | Affinity Capture-Western | BioGRID | 9535896 |11712088 |
| DKK1 | DKK-1 | SK | dickkopf homolog 1 (Xenopus laevis) | CTNNB1 (beta-catenin) interacts with the DKK1 promoter. | BIND | 15592430 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | - | HPRD | 9512503 |
| DLG5 | KIAA0583 | LP-DLG | P-DLG5 | PDLG | discs, large homolog 5 (Drosophila) | - | HPRD,BioGRID | 12657639 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | Affinity Capture-Western | BioGRID | 9286858 |
| DSC3 | CDHF3 | DSC | DSC1 | DSC2 | DSC4 | HT-CP | desmocollin 3 | - | HPRD,BioGRID | 10769211 |
| DVL1 | DVL | MGC54245 | dishevelled, dsh homolog 1 (Drosophila) | - | HPRD,BioGRID | 10806215 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Affinity Capture-Western | BioGRID | 10806215 |
| DVL3 | KIAA0208 | dishevelled, dsh homolog 3 (Drosophila) | - | HPRD,BioGRID | 10806215 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 9233779 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD,BioGRID | 7702605 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Affinity Capture-Western | BioGRID | 15304487 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | beta-catenin interacts with ER-alpha. | BIND | 15304487 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | - | HPRD,BioGRID | 10462524 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | CTNNB1 (beta-catenin) interacts with the FAS promoter. | BIND | 15829968 |
| FER | TYK3 | fer (fps/fes related) tyrosine kinase | Biochemical Activity | BioGRID | 12640114 |
| FGF20 | - | fibroblast growth factor 20 | CTNNB1 (beta-catenin) interacts with the FGF20 promoter. | BIND | 15592430 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | - | HPRD,BioGRID | 12466281 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | beta-catenin interacts with FOXO1. This interaction was modelled on a demonstrated interaction between beta-catenin from an unspecified species and human FOXO1. | BIND | 15905404 |
| FOXO3 | AF6q21 | DKFZp781A0677 | FKHRL1 | FKHRL1P2 | FOXO2 | FOXO3A | MGC12739 | MGC31925 | forkhead box O3 | beta-catenin interacts with FOXO3a. | BIND | 15905404 |
| FOXO4 | AFX | AFX1 | MGC120490 | MLLT7 | forkhead box O4 | beta-catenin interacts with FOXO4. This interaction was modelled on a demonstrated interaction between human FOXO4 and beta-catenin from an unspecified species. | BIND | 15905404 |
| FSCN2 | RFSN | RP30 | fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) | - | HPRD | 11795941 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | Biochemical Activity | BioGRID | 12640114 |
| GNA13 | G13 | MGC46138 | guanine nucleotide binding protein (G protein), alpha 13 | - | HPRD | 11136230 |
| GRIK2 | EAA4 | GLR6 | GLUK6 | GLUR6 | MGC74427 | MRT6 | glutamate receptor, ionotropic, kainate 2 | - | HPRD | 12151522 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| GSK3B | - | glycogen synthase kinase 3 beta | GSK3B (GSK3-Beta) interacts with and phosphorylates CTNNB1 (Beta-catenin). | BIND | 15829978 |
| GSK3B | - | glycogen synthase kinase 3 beta | Affinity Capture-Western Biochemical Activity | BioGRID | 10330181 |11251183 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | - | HPRD | 11266540 |12107263 |
| HNF4A | FLJ39654 | HNF4 | HNF4a7 | HNF4a8 | HNF4a9 | MODY | MODY1 | NR2A1 | NR2A21 | TCF | TCF14 | hepatocyte nuclear factor 4, alpha | Affinity Capture-Western Reconstituted Complex | BioGRID | 12944908 |
| IGFBP2 | IBP2 | IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa | CTNNB1 (beta-catenin) interacts with the IGFBP2 promoter. | BIND | 15735679 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | - | HPRD,BioGRID | 11527961 |
| IQGAP1 | HUMORFA01 | KIAA0051 | SAR1 | p195 | IQ motif containing GTPase activating protein 1 | - | HPRD,BioGRID | 11734550|12446675 |
| JUP | ARVD12 | CTNNG | DP3 | DPIII | PDGB | PKGB | junction plakoglobin | Affinity Capture-Western | BioGRID | 9535896 |11712088 |
| KLK3 | APS | KLK2A1 | PSA | hK3 | kallikrein-related peptidase 3 | CTNNB1 (beta-catenin) interacts with the KLK3 (PSA) promoter. | BIND | 15829968 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | - | HPRD | 8757136 |12556497 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | Affinity Capture-Western Two-hybrid | BioGRID | 8757136 |12748295 |15684397 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | LEF-1 interacts with Beta-catenin. This interaction was modeled on a demonstrated interaction between human LEF-1 and mouse Beta-catenin. | BIND | 15525529 |
| LGALS9 | HUAT | LGALS9A | MGC117375 | MGC125973 | MGC125974 | lectin, galactoside-binding, soluble, 9 | Affinity Capture-MS | BioGRID | 17353931 |
| MAGI1 | AIP3 | BAIAP1 | BAP1 | MAGI-1 | TNRC19 | WWP3 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10772923 |
| MAGI2 | ACVRIP1 | AIP1 | ARIP1 | MAGI-2 | SSCAM | membrane associated guanylate kinase, WW and PDZ domain containing 2 | - | HPRD | 11826105 |
| MET | AUTS9 | HGFR | RCCP2 | c-Met | met proto-oncogene (hepatocyte growth factor receptor) | Affinity Capture-Western | BioGRID | 11254878 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | Affinity Capture-Western Reconstituted Complex | BioGRID | 9139698 |9819408 |11152665 |11483589 |11877440 |12618757 |12750561 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | MUC1 interacts with beta-catenin. | BIND | 11483589 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | - | HPRD | 9139698 |11152665 |11483589 |11877440 |
| MYO7A | DFNA11 | DFNB2 | MYOVIIA | MYU7A | NSRD2 | USH1B | myosin VIIA | Affinity Capture-Western | BioGRID | 11080149 |
| NEURL2 | C20orf163 | MGC125934 | MGC125935 | OZZ | Ozz-E3 | neuralized homolog 2 (Drosophila) | - | HPRD | 14960280 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | NFKB1 (p50) interacts with CTNNB1 (beta-catenin). | BIND | 15829968 |
| NLK | DKFZp761G1211 | FLJ21033 | nemo-like kinase | Affinity Capture-Western | BioGRID | 12556497 |
| NR5A1 | AD4BP | ELP | FTZ1 | FTZF1 | SF-1 | SF1 | nuclear receptor subfamily 5, group A, member 1 | - | HPRD,BioGRID | 12554773 |
| NR5A2 | B1F | B1F2 | CPF | FTF | FTZ-F1 | FTZ-F1beta | LRH-1 | hB1F | hB1F-2 | nuclear receptor subfamily 5, group A, member 2 | beta-catenin interacts with LRH-1. | BIND | 15327767 |
| PCSK1 | BMIQ12 | NEC1 | PC1 | PC3 | SPC3 | proprotein convertase subtilisin/kexin type 1 | Affinity Capture-Western | BioGRID | 11113628 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | - | HPRD,BioGRID | 10801826 |
| PHB2 | BAP | BCAP37 | Bap37 | MGC117268 | PNAS-141 | REA | p22 | prohibitin 2 | Affinity Capture-MS | BioGRID | 17353931 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Reconstituted Complex | BioGRID | 10477752 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD | 11716761 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | - | HPRD,BioGRID | 11533658 |
| PKD1 | PBP | polycystic kidney disease 1 (autosomal dominant) | Polycystin-1 interacts with beta-catenin. | BIND | 11113628 |
| PKD1 | PBP | polycystic kidney disease 1 (autosomal dominant) | - | HPRD | 11274246 |
| PKP2 | ARVD9 | plakophilin 2 | - | HPRD | 11790773 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | Affinity Capture-Western | BioGRID | 9738936 |9852041 |10341227 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | PS1 interacts with beta-catenin. | BIND | 9632714 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | Affinity Capture-Western | BioGRID | 10706604 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD,BioGRID | 12377785 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Affinity Capture-Western | BioGRID | 10681592 |
| PTPN13 | DKFZp686J1497 | FAP-1 | PNP1 | PTP-BAS | PTP-BL | PTP1E | PTPL1 | PTPLE | protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) | Reconstituted Complex | BioGRID | 10951583 |
| PTPN14 | MGC126803 | PEZ | PTP36 | protein tyrosine phosphatase, non-receptor type 14 | - | HPRD,BioGRID | 12808048 |
| PTPRB | DKFZp686E2262 | DKFZp686H15164 | FLJ44133 | HPTP-BETA | HPTPB | MGC142023 | MGC59935 | PTPB | R-PTP-BETA | VEPTP | protein tyrosine phosphatase, receptor type, B | - | HPRD | 10706604 |
| PTPRF | FLJ43335 | FLJ45062 | FLJ45567 | LAR | protein tyrosine phosphatase, receptor type, F | Affinity Capture-Western Reconstituted Complex | BioGRID | 9245795 |11245482 |
| PTPRF | FLJ43335 | FLJ45062 | FLJ45567 | LAR | protein tyrosine phosphatase, receptor type, F | - | HPRD | 9245795 |12095414 |
| PTPRJ | CD148 | DEP1 | HPTPeta | R-PTP-ETA | SCC1 | protein tyrosine phosphatase, receptor type, J | - | HPRD,BioGRID | 12370829 |
| PTPRK | DKFZp686C2268 | DKFZp779N1045 | R-PTP-kappa | protein tyrosine phosphatase, receptor type, K | - | HPRD,BioGRID | 8663237 |
| PTPRU | FLJ37530 | FMI | GLEPP1 | PCP-2 | PTP | PTP-J | PTP-PI | PTPPSI | PTPRO | PTPU2 | R-PTP-PSI | hPTP-J | protein tyrosine phosphatase, receptor type, U | - | HPRD,BioGRID | 12501215 |
| PTPRZ1 | HPTPZ | HPTPzeta | PTP-ZETA | PTP18 | PTPRZ | PTPZ | RPTPB | RPTPbeta | phosphacan | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | - | HPRD,BioGRID | 10706604 |
| PYGO1 | DKFZp547G0910 | pygopus homolog 1 (Drosophila) | - | HPRD | 11955446 |
| RAB8B | FLJ38125 | RAB8B, member RAS oncogene family | Affinity Capture-Western | BioGRID | 12639940 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | - | HPRD | 12531901 |
| RUVBL1 | ECP54 | INO80H | NMP238 | PONTIN | Pontin52 | RVB1 | TIH1 | TIP49 | TIP49A | RuvB-like 1 (E. coli) | Affinity Capture-Western Reconstituted Complex | BioGRID | 9843967 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | CTNNB1 (beta-catenin) interacts with RUVBL2 (reptin). | BIND | 15829968 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | Affinity Capture-Western | BioGRID | 12771132 |
| SGK1 | SGK | serum/glucocorticoid regulated kinase 1 | CTNNB1 (beta-catenin) interacts with the SGK promoter. | BIND | 15735679 |
| SKP1 | EMC19 | MGC34403 | OCP-II | OCP2 | SKP1A | TCEB1L | p19A | S-phase kinase-associated protein 1 | Affinity Capture-Western | BioGRID | 11389839 |
| SLC9A3R1 | EBP50 | NHERF | NHERF1 | NPHLOP2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | - | HPRD,BioGRID | 12830000 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 12000714 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 12000714 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | - | HPRD,BioGRID | 12000714 |
| SMAD7 | CRCS3 | FLJ16482 | MADH7 | MADH8 | SMAD family member 7 | Affinity Capture-Western Co-localization | BioGRID | 15684397 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | - | HPRD,BioGRID | 11532957 |
| TAX1BP3 | TIP-1 | Tax1 (human T-cell leukemia virus type I) binding protein 3 | - | HPRD | 12874278 |
| TBL1X | EBI | SMAP55 | TBL1 | transducin (beta)-like 1X-linked | - | HPRD,BioGRID | 11389839 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | - | HPRD | 12107263 |
| TCF4 | E2-2 | ITF2 | MGC149723 | MGC149724 | PTHS | SEF2 | SEF2-1 | SEF2-1A | SEF2-1B | bHLHb19 | transcription factor 4 | - | HPRD | 11713476|12861022 |
| TCF4 | E2-2 | ITF2 | MGC149723 | MGC149724 | PTHS | SEF2 | SEF2-1 | SEF2-1A | SEF2-1B | bHLHb19 | transcription factor 4 | Affinity Capture-Western Co-crystal Structure Reconstituted Complex | BioGRID | 11713475 |11713476 |12657632 |15331612 |
| TCF7L1 | TCF-3 | TCF3 | transcription factor 7-like 1 (T-cell specific, HMG-box) | Co-crystal Structure | BioGRID | 11136974 |
| TCF7L1 | TCF-3 | TCF3 | transcription factor 7-like 1 (T-cell specific, HMG-box) | - | HPRD | 11266540 |
| TCF7L2 | TCF-4 | TCF4 | transcription factor 7-like 2 (T-cell specific, HMG-box) | Affinity Capture-Western | BioGRID | 12861022 |
| TCF7L2 | TCF-4 | TCF4 | transcription factor 7-like 2 (T-cell specific, HMG-box) | - | HPRD | 11713476 |
| TCF7L2 | TCF-4 | TCF4 | transcription factor 7-like 2 (T-cell specific, HMG-box) | TCF-4 interacts with Beta-catenin. | BIND | 15525529 |
| TCF7L2 | TCF-4 | TCF4 | transcription factor 7-like 2 (T-cell specific, HMG-box) | Beta-catenin interacts with TCF4. | BIND | 15294866 |
| TFAP2A | AP-2 | AP-2alpha | AP2TF | BOFS | TFAP2 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | Affinity Capture-Western | BioGRID | 15331612 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | beta-catenin interacts with the pS2 promoter. | BIND | 15304487 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | - | HPRD | 12000714 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | Affinity Capture-Western | BioGRID | 12000714 |
| USP9X | DFFRX | FAF | FAM | ubiquitin specific peptidase 9, X-linked | - | HPRD,BioGRID | 10620020 |
| VCL | CMD1W | MVCL | vinculin | Affinity Capture-Western | BioGRID | 9405455 |
| VEZT | DKFZp761C241 | VEZATIN | vezatin, adherens junctions transmembrane protein | - | HPRD,BioGRID | 11080149 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER | 29 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELL2CELL PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GSK3 PATHWAY | 27 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PITX2 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PS1 PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TFF PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| ST WNT BETA CATENIN PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN DEG PATHWAY | 18 | 17 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID NECTIN PATHWAY | 30 | 20 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID WNT CANONICAL PATHWAY | 20 | 18 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN NASCENT AJ PATHWAY | 39 | 33 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN KERATINOCYTE PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID NCADHERIN PATHWAY | 33 | 32 | All SZGR 2.0 genes in this pathway |
| PID RAC1 PATHWAY | 54 | 37 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY WNT | 65 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME CTNNB1 PHOSPHORYLATION CASCADE | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | 12 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME ADHERENS JUNCTIONS INTERACTIONS | 27 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME MYOGENESIS | 28 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | 22 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| KOINUMA COLON CANCER MSI UP | 16 | 11 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HWANG PROSTATE CANCER MARKERS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK TARGETS | 18 | 15 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA DN | 28 | 21 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS UP | 79 | 46 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C5 | 21 | 11 | All SZGR 2.0 genes in this pathway |
| MA PITUITARY FETAL VS ADULT UP | 29 | 21 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA DELAYED RESPONSE TO TGFB1 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 30MIN | 54 | 36 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH UP | 74 | 56 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 | 71 | 42 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| HONMA DOCETAXEL RESISTANCE | 34 | 23 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| ZAIDI OSTEOBLAST TRANSCRIPTION FACTORS | 14 | 12 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| GUILLAUMOND KLF10 TARGETS DN | 30 | 15 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-139 | 349 | 355 | 1A | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-141/200a | 697 | 703 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-217 | 669 | 675 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-320 | 239 | 245 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-381 | 829 | 835 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.