Gene Page: DBH
Summary ?
| GeneID | 1621 |
| Symbol | DBH |
| Synonyms | DBM |
| Description | dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| Reference | MIM:609312|HGNC:HGNC:2689|Ensembl:ENSG00000123454|HPRD:01963|Vega:OTTHUMG00000020878 |
| Gene type | protein-coding |
| Map location | 9q34 |
| Pascal p-value | 0.089 |
| Fetal beta | 0.137 |
| eGene | Cortex Frontal Cortex BA9 Meta |
| Support | NEUROTRANSMITTER METABOLISM |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
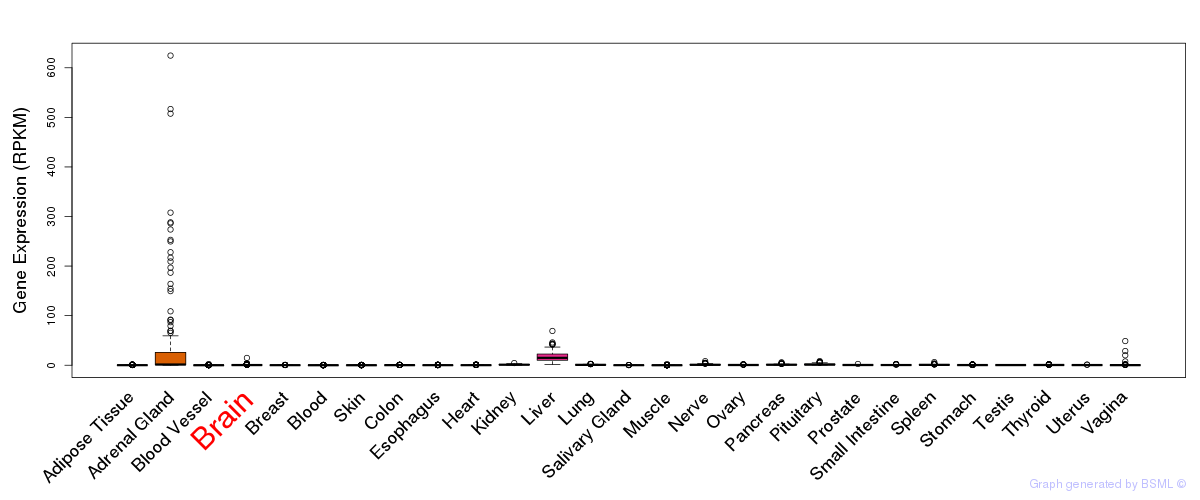
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PPP1CB | 0.92 | 0.91 |
| FAM49B | 0.91 | 0.90 |
| STX12 | 0.90 | 0.91 |
| RAB2A | 0.90 | 0.91 |
| CNOT7 | 0.90 | 0.89 |
| UBE2N | 0.90 | 0.88 |
| AZIN1 | 0.90 | 0.88 |
| TMEM68 | 0.90 | 0.89 |
| RAB18 | 0.90 | 0.90 |
| TTC33 | 0.89 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.65 | -0.64 |
| AF347015.8 | -0.65 | -0.66 |
| AF347015.26 | -0.65 | -0.65 |
| AF347015.2 | -0.65 | -0.64 |
| MT-CYB | -0.64 | -0.64 |
| AF347015.33 | -0.64 | -0.63 |
| AF347015.15 | -0.63 | -0.64 |
| AF347015.31 | -0.61 | -0.61 |
| GPT | -0.60 | -0.63 |
| AF347015.27 | -0.60 | -0.61 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004500 | dopamine beta-monooxygenase activity | IEA | dopamine (GO term level: 6) | - |
| GO:0005507 | copper ion binding | IEA | - | |
| GO:0004497 | monooxygenase activity | IEA | - | |
| GO:0031418 | L-ascorbic acid binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 2880016 |
| GO:0009987 | cellular process | IEA | - | |
| GO:0006548 | histidine catabolic process | IEA | - | |
| GO:0042423 | catecholamine biosynthetic process | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005624 | membrane fraction | TAS | 3443096 | |
| GO:0005625 | soluble fraction | TAS | 3443096 | |
| GO:0005737 | cytoplasm | TAS | 3443096 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0042583 | chromaffin granule | NAS | - | |
| GO:0031410 | cytoplasmic vesicle | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TYROSINE METABOLISM | 42 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NEUROTRANSMITTERS PATHWAY | 6 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE DERIVED HORMONES | 15 | 12 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| RORIE TARGETS OF EWSR1 FLI1 FUSION DN | 30 | 23 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS UP | 93 | 56 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |