Gene Page: EDA
Summary ?
| GeneID | 1896 |
| Symbol | EDA |
| Synonyms | ECTD1|ED1|ED1-A1|ED1-A2|EDA-A1|EDA-A2|EDA1|EDA2|HED|HED1|ODT1|STHAGX1|TNLG7C|XHED|XLHED |
| Description | ectodysplasin A |
| Reference | MIM:300451|HGNC:HGNC:3157|Ensembl:ENSG00000158813|HPRD:02347|Vega:OTTHUMG00000021764 |
| Gene type | protein-coding |
| Map location | Xq12-q13.1 |
| Fetal beta | 1.25 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
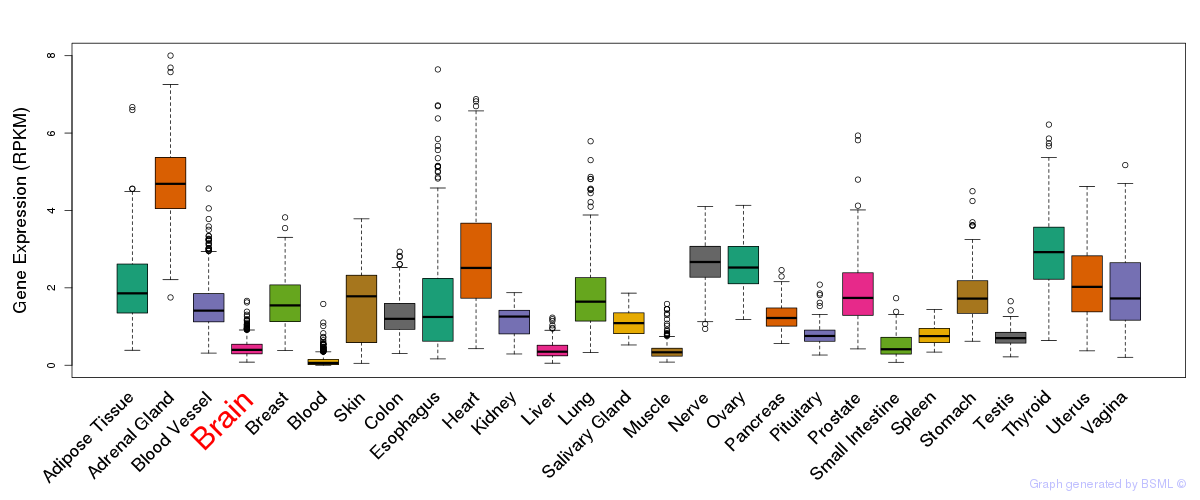
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LAMA4 | 0.76 | 0.73 |
| ETS1 | 0.73 | 0.78 |
| LAMC1 | 0.71 | 0.70 |
| FN1 | 0.71 | 0.71 |
| VCL | 0.69 | 0.70 |
| PCDH18 | 0.69 | 0.70 |
| FLVCR2 | 0.69 | 0.59 |
| NID1 | 0.69 | 0.64 |
| ELK3 | 0.68 | 0.72 |
| OLFML3 | 0.68 | 0.63 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.39 | -0.36 |
| AF347015.21 | -0.39 | -0.38 |
| MT-CO2 | -0.37 | -0.40 |
| S100A13 | -0.36 | -0.34 |
| MT1G | -0.36 | -0.34 |
| AC098691.2 | -0.35 | -0.36 |
| APOC1 | -0.34 | -0.30 |
| AF347015.31 | -0.34 | -0.37 |
| CA5A | -0.34 | -0.32 |
| AF347015.8 | -0.34 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IDA | Neurotransmitter (GO term level: 4) | 11039935 |
| GO:0005164 | tumor necrosis factor receptor binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001942 | hair follicle development | IEA | - | |
| GO:0007160 | cell-matrix adhesion | IEA | - | |
| GO:0007398 | ectoderm development | TAS | 8696334 | |
| GO:0007165 | signal transduction | TAS | 8696334 | |
| GO:0007431 | salivary gland development | IEA | - | |
| GO:0006955 | immune response | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0042346 | positive regulation of NF-kappaB import into nucleus | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | IDA | 11039935 | |
| GO:0042475 | odontogenesis of dentine-containing tooth | IEA | - | |
| GO:0043473 | pigmentation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | TAS | 10484778 | |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005624 | membrane fraction | TAS | 8696334 | |
| GO:0005886 | plasma membrane | TAS | 9736768 |10484778 | |
| GO:0005887 | integral to plasma membrane | IEA | - | |
| GO:0045177 | apical part of cell | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| WANG PROSTATE CANCER ANDROGEN INDEPENDENT | 66 | 37 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 845 | 851 | m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-130/301 | 313 | 320 | 1A,m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-149 | 46 | 52 | m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-15/16/195/424/497 | 72 | 78 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-150 | 42 | 48 | m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-22 | 627 | 633 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-24 | 3039 | 3046 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-324-5p | 3142 | 3148 | 1A | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-342 | 3748 | 3755 | 1A,m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.