Gene Page: AHR
Summary ?
| GeneID | 196 |
| Symbol | AHR |
| Synonyms | bHLHe76 |
| Description | aryl hydrocarbon receptor |
| Reference | MIM:600253|HGNC:HGNC:348|Ensembl:ENSG00000106546|HPRD:02596|Vega:OTTHUMG00000149967 |
| Gene type | protein-coding |
| Map location | 7p15 |
| Pascal p-value | 0.609 |
| Sherlock p-value | 0.817 |
| Fetal beta | -0.8 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
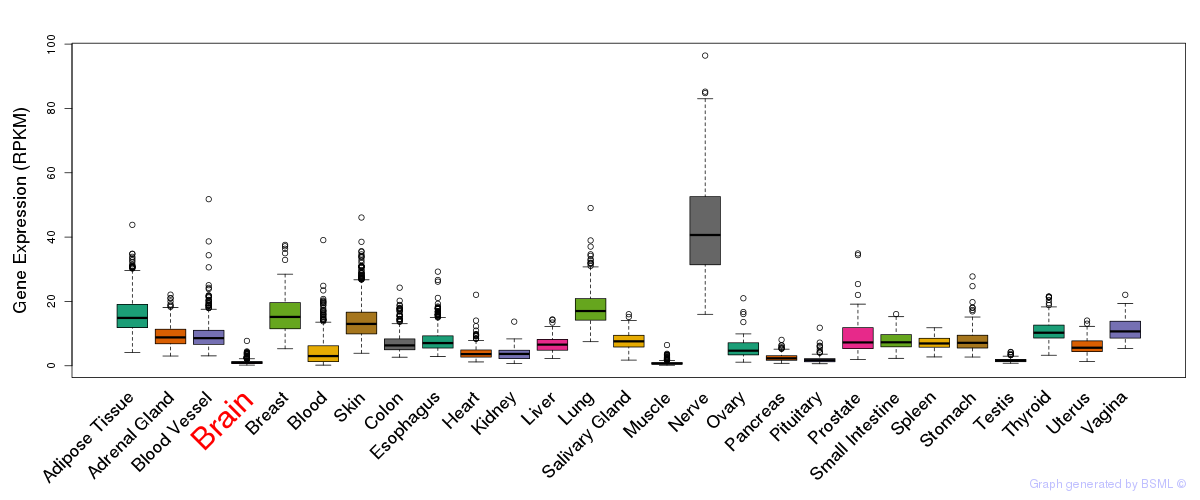
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RGS10 | 0.88 | 0.90 |
| C3 | 0.82 | 0.82 |
| LY86 | 0.81 | 0.83 |
| SAMSN1 | 0.79 | 0.79 |
| TREM2 | 0.78 | 0.79 |
| GPR34 | 0.77 | 0.84 |
| CD86 | 0.76 | 0.79 |
| ADORA3 | 0.76 | 0.77 |
| LAPTM5 | 0.75 | 0.75 |
| PGDS | 0.75 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SCUBE1 | -0.36 | -0.32 |
| SMTN | -0.36 | -0.47 |
| SRGAP1 | -0.33 | -0.22 |
| GJC1 | -0.33 | -0.31 |
| HIP1R | -0.33 | -0.37 |
| FBN3 | -0.33 | -0.27 |
| PNPLA3 | -0.33 | -0.24 |
| SEMA4B | -0.33 | -0.42 |
| NAV2 | -0.33 | -0.22 |
| MN1 | -0.32 | -0.17 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AIP | ARA9 | FKBP16 | FKBP37 | SMTPHN | XAP2 | aryl hydrocarbon receptor interacting protein | - | HPRD,BioGRID | 7961671 |9083006 |9111057 |9837941 |10986286 |12065584 |
| AIP | ARA9 | FKBP16 | FKBP37 | SMTPHN | XAP2 | aryl hydrocarbon receptor interacting protein | - | HPRD | 9111057 |9837941 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | - | HPRD | 7488247 |7628454 |8384309 |11768231 |12024042 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | Arnt interacts with AhR. | BIND | 9704006 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 7488247 |7628454 |8384309 |9887096 |
| ARNTL | BMAL1 | BMAL1c | JAP3 | MGC47515 | MOP3 | PASD3 | TIC | bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like | - | HPRD,BioGRID | 9079689 |
| CCNT1 | CCNT | CYCT1 | cyclin T1 | - | HPRD,BioGRID | 12917420 |
| DAP3 | DAP-3 | DKFZp686G12159 | MGC126058 | MGC126059 | MRP-S29 | MRPS29 | bMRP-10 | death associated protein 3 | - | HPRD,BioGRID | 10903152 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 10620335 |12612060 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Reconstituted Complex | BioGRID | 8794892 |
| GTF2F2 | BTF4 | RAP30 | TF2F2 | TFIIF | general transcription factor IIF, polypeptide 2, 30kDa | - | HPRD,BioGRID | 8794892 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 7961671 |9083006 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD | 12024042 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | AHR interacts with NCOA1 (SRC-1a). | BIND | 15641800 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | Affinity Capture-Western | BioGRID | 12024042 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | AHR interacts with NCOA2 (TIF2). | BIND | 15641800 |
| NEDD8 | FLJ43224 | MGC104393 | MGC125896 | MGC125897 | Nedd-8 | neural precursor cell expressed, developmentally down-regulated 8 | - | HPRD,BioGRID | 12215427 |
| NR2F1 | COUP-TFI | EAR-3 | EAR3 | ERBAL3 | NR2F2 | SVP44 | TCFCOUP1 | TFCOUP1 | nuclear receptor subfamily 2, group F, member 1 | - | HPRD,BioGRID | 10620335 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | - | HPRD,BioGRID | 10428779 |
| PTGES3 | P23 | TEBP | cPGES | prostaglandin E synthase 3 (cytosolic) | - | HPRD | 11013261 |11259606 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 9712901 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 11114727 |12181450 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | - | HPRD,BioGRID | 11805098 |
| SP1 | - | Sp1 transcription factor | - | HPRD,BioGRID | 10471301 |12612060 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12024042 |
| TAF4 | FLJ41943 | TAF2C | TAF2C1 | TAF4A | TAFII130 | TAFII135 | TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa | AHR interacts with TAF4. | BIND | 15641800 |
| TAF6 | DKFZp781E21155 | MGC:8964 | TAF2E | TAFII70 | TAFII80 | TAFII85 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa | AHR interacts with TAF6. | BIND | 15641800 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | AHR interacts with TBP. | BIND | 15641800 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 8794892 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | - | HPRD | 12065584 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| SHI SPARC TARGETS DN | 13 | 8 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| FRIDMAN IMMORTALIZATION DN | 34 | 24 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR DN | 41 | 30 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 11Q23 DELETION | 23 | 18 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN | 56 | 37 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN | 101 | 70 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 11 | 57 | 40 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| MATZUK EARLY ANTRAL FOLLICLE | 13 | 11 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS UP | 14 | 11 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 9 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PLASARI NFIC TARGETS BASAL DN | 18 | 13 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR UP | 33 | 25 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR DN | 30 | 25 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |