Gene Page: FANCG
Summary ?
| GeneID | 2189 |
| Symbol | FANCG |
| Synonyms | FAG|XRCC9 |
| Description | Fanconi anemia complementation group G |
| Reference | MIM:602956|HGNC:HGNC:3588|Ensembl:ENSG00000221829|HPRD:04262|Vega:OTTHUMG00000019850 |
| Gene type | protein-coding |
| Map location | 9p13 |
| Pascal p-value | 0.008 |
| Sherlock p-value | 0.331 |
| Fetal beta | 0.87 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03563403 | 9 | 35079882 | FANCG | 9.47E-11 | -0.01 | 4.49E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | FANCG | 2189 | 1.658E-8 | trans | ||
| rs17762315 | chr5 | 76807576 | FANCG | 2189 | 0.02 | trans | ||
| rs16955618 | chr15 | 29937543 | FANCG | 2189 | 3.699E-7 | trans |
Section II. Transcriptome annotation
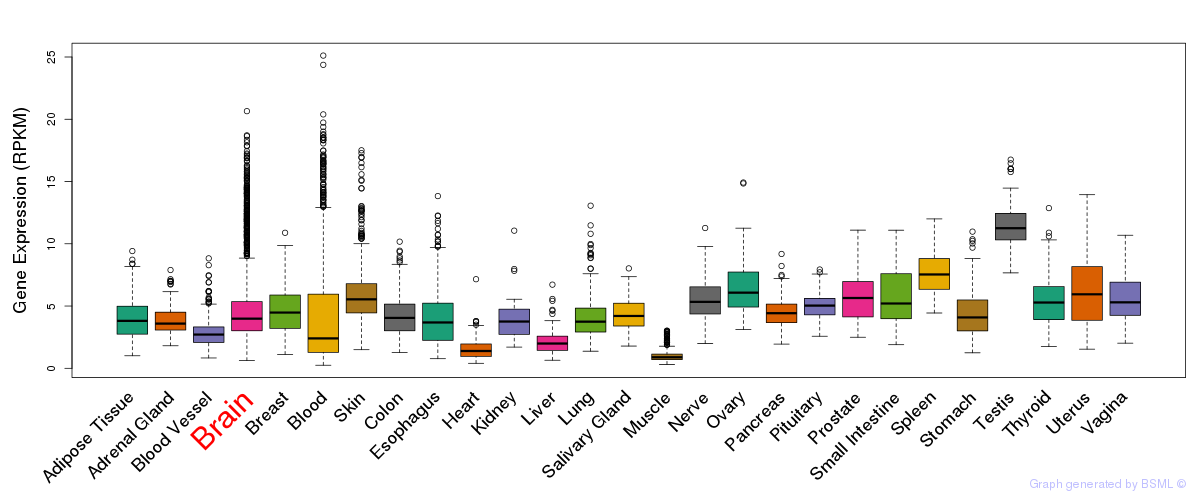
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | Affinity Capture-Western Two-hybrid | BioGRID | 12915460 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | FANCG interacts with BRCA2. | BIND | 12915460 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | Reconstituted Complex | BioGRID | 15367677 |
| CYP19A1 | ARO | ARO1 | CPV1 | CYAR | CYP19 | MGC104309 | P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 | - | HPRD | 11756225 |
| CYP2E1 | CPE1 | CYP2E | P450-J | P450C2E | cytochrome P450, family 2, subfamily E, polypeptide 1 | Two-hybrid | BioGRID | 14499622 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | FANCA interacts with FANCG. | BIND | 15138265 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | - | HPRD | 10468603 |11050007 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Affinity Capture-MS Affinity Capture-Western Co-purification in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 10373536 |10468603 |10468606 |10567393 |10627486 |10652215 |10961856 |11050007 |11157805 |11739169 |11918676 |12210728 |12239156 |12649160 |12973351 |14697762 |15082718 |15138265 |16189514 |
| FANCC | FA3 | FAC | FACC | FLJ14675 | Fanconi anemia, complementation group C | - | HPRD,BioGRID | 10373536 |
| FANCE | FACE | FAE | Fanconi anemia, complementation group E | - | HPRD | 11157805 |12239156 |
| FANCE | FACE | FAE | Fanconi anemia, complementation group E | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 11157805 |12093742 |12239156 |
| FANCF | FAF | MGC126856 | Fanconi anemia, complementation group F | - | HPRD,BioGRID | 11063725 |11157805 |
| FANCG | FAG | XRCC9 | Fanconi anemia, complementation group G | FANCG interacts with FANCG. | BIND | 15138265 |
| FANCG | FAG | XRCC9 | Fanconi anemia, complementation group G | Co-purification Two-hybrid | BioGRID | 10652215 |11157805 |15138265 |
| GABPB1 | BABPB2 | E4TF1 | E4TF1-47 | E4TF1-53 | E4TF1B | GABPB | GABPB2 | NRF2B1 | NRF2B2 | GA binding protein transcription factor, beta subunit 1 | Two-hybrid | BioGRID | 16189514 |
| KRT19 | CK19 | K19 | K1CS | MGC15366 | keratin 19 | Two-hybrid | BioGRID | 16189514 |
| PSMB1 | FLJ25321 | HC5 | KIAA1838 | PMSB1 | PSC5 | proteasome (prosome, macropain) subunit, beta type, 1 | Two-hybrid | BioGRID | 14499622 |
| RTN2 | NSP2 | NSPL1 | reticulon 2 | Two-hybrid | BioGRID | 14499622 |
| SAMD3 | FLJ34563 | MGC35163 | sterile alpha motif domain containing 3 | Two-hybrid | BioGRID | 16189514 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | Co-purification | BioGRID | 11401546 |
| USHBP1 | AIEBP | FLJ38709 | FLJ90681 | MCC2 | Usher syndrome 1C binding protein 1 | Two-hybrid | BioGRID | 16189514 |
| ZBED1 | ALTE | DREF | KIAA0785 | TRAMP | hDREF | zinc finger, BED-type containing 1 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA ATRBRCA PATHWAY | 21 | 12 | All SZGR 2.0 genes in this pathway |
| PID FANCONI PATHWAY | 47 | 28 | All SZGR 2.0 genes in this pathway |
| PID BARD1 PATHWAY | 29 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME FANCONI ANEMIA PATHWAY | 25 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS UP | 27 | 17 | All SZGR 2.0 genes in this pathway |
| WAKASUGI HAVE ZNF143 BINDING SITES | 58 | 33 | All SZGR 2.0 genes in this pathway |
| MATHEW FANCONI ANEMIA GENES | 11 | 7 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN MELANOMA RELAPSE UP | 61 | 25 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 8 | 36 | 28 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL FLI1 | 9 | 8 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| MATZUK MEIOTIC AND DNA REPAIR | 39 | 26 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| CHANG CYCLING GENES | 148 | 83 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G1 S | 147 | 76 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |