Gene Page: MYO16
Summary ?
| GeneID | 23026 |
| Symbol | MYO16 |
| Synonyms | MYAP3|MYR8|Myo16b|NYAP3|PPP1R107 |
| Description | myosin XVI |
| Reference | MIM:615479|HGNC:HGNC:29822|Ensembl:ENSG00000041515|HPRD:14795|Vega:OTTHUMG00000017333 |
| Gene type | protein-coding |
| Map location | 13q33.3 |
| Pascal p-value | 0.026 |
| Sherlock p-value | 0.355 |
| DMG | 1 (# studies) |
| Support | CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26899526 | 13 | 109794149 | MYO16 | 8.36E-5 | 0.459 | 0.026 | DMG:Wockner_2014 |
| cg24425807 | 13 | 109741176 | MYO16 | 3.71E-4 | 0.699 | 0.043 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
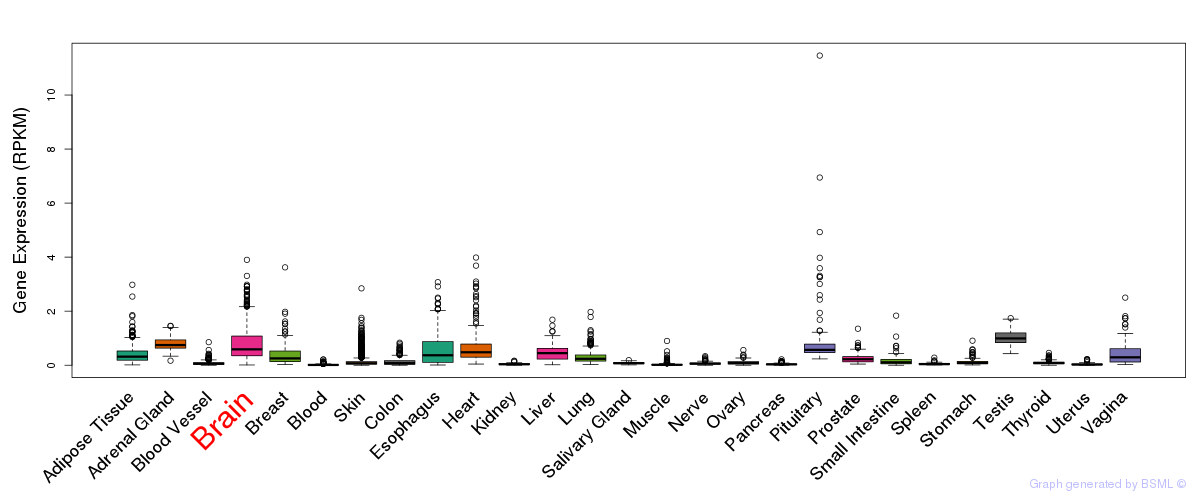
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1219 | 0.95 | 0.96 |
| HECTD1 | 0.95 | 0.96 |
| FAM160B1 | 0.94 | 0.96 |
| RNMT | 0.94 | 0.95 |
| ZBTB38 | 0.93 | 0.94 |
| USP9X | 0.93 | 0.95 |
| GNAQ | 0.93 | 0.96 |
| BAG5 | 0.93 | 0.95 |
| CPEB4 | 0.93 | 0.95 |
| ZYG11B | 0.93 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.71 | -0.76 |
| MT-CO2 | -0.69 | -0.76 |
| HIGD1B | -0.68 | -0.76 |
| AF347015.31 | -0.68 | -0.74 |
| HSD17B14 | -0.66 | -0.71 |
| TLCD1 | -0.66 | -0.71 |
| ENHO | -0.66 | -0.76 |
| AC021016.1 | -0.65 | -0.70 |
| IFI27 | -0.65 | -0.72 |
| AF347015.33 | -0.65 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003774 | motor activity | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0051015 | actin filament binding | ISS | 11588169 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0021549 | cerebellum development | ISS | Brain (GO term level: 10) | 11588169 |
| GO:0008285 | negative regulation of cell proliferation | ISS | 17029291 | |
| GO:0045749 | negative regulation of S phase of mitotic cell cycle | ISS | 17029291 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005654 | nucleoplasm | IDA | 17029291 | |
| GO:0005737 | cytoplasm | ISS | 11588169 | |
| GO:0005886 | plasma membrane | ISS | 11588169 |17029291 | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 17029291 | |
| GO:0016459 | myosin complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |