Gene Page: CIC
Summary ?
| GeneID | 23152 |
| Symbol | CIC |
| Synonyms | - |
| Description | capicua transcriptional repressor |
| Reference | MIM:612082|HGNC:HGNC:14214|Ensembl:ENSG00000079432|HPRD:10831|Vega:OTTHUMG00000182794 |
| Gene type | protein-coding |
| Map location | 19q13.2 |
| Pascal p-value | 0.125 |
| Sherlock p-value | 0.19 |
| TADA p-value | 0.027 |
| Fetal beta | 0.964 |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0505 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CIC | chr19 | 42791987 | A | G | NM_015125 | p.264H>R | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | CIC | 23152 | 1.779E-4 | trans | ||
| rs3845734 | chr2 | 171125572 | CIC | 23152 | 0.18 | trans | ||
| rs7787830 | chr7 | 98797019 | CIC | 23152 | 0.18 | trans | ||
| rs16955618 | chr15 | 29937543 | CIC | 23152 | 0 | trans |
Section II. Transcriptome annotation
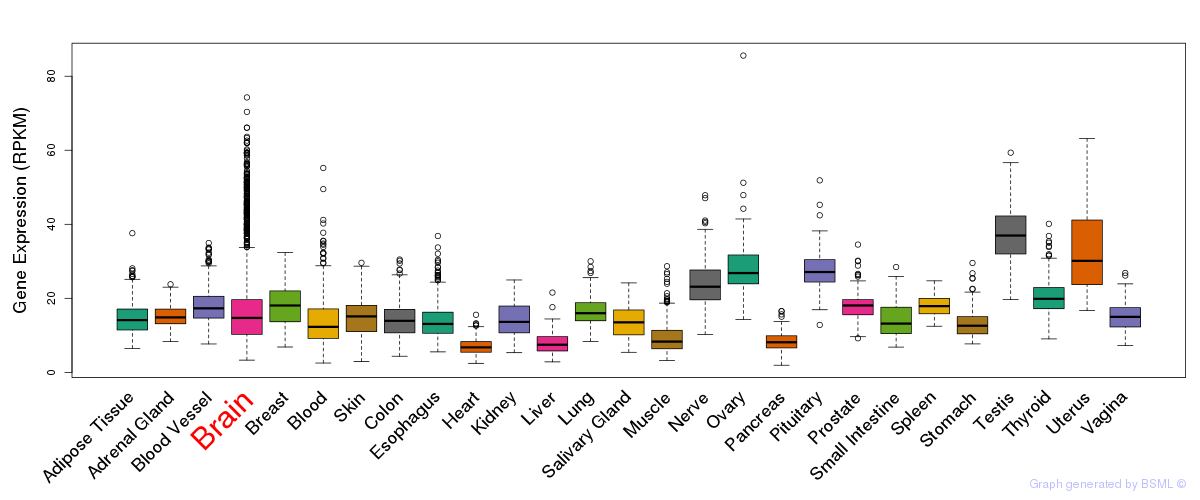
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GTSE1 | 0.99 | 0.84 |
| KIF2C | 0.98 | 0.79 |
| NUSAP1 | 0.98 | 0.81 |
| CDCA8 | 0.98 | 0.76 |
| KIF20A | 0.98 | 0.80 |
| TACC3 | 0.98 | 0.73 |
| ORC1L | 0.98 | 0.77 |
| GSG2 | 0.98 | 0.79 |
| KIF4A | 0.98 | 0.81 |
| CDK2 | 0.98 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.39 | -0.69 |
| FBXO2 | -0.39 | -0.65 |
| HLA-F | -0.39 | -0.72 |
| PTH1R | -0.39 | -0.69 |
| SLC9A3R2 | -0.38 | -0.46 |
| ALDOC | -0.38 | -0.69 |
| AIFM3 | -0.38 | -0.70 |
| ASPHD1 | -0.37 | -0.53 |
| AF347015.27 | -0.37 | -0.80 |
| LHPP | -0.37 | -0.57 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 16713569 | |
| GO:0005215 | transporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| DASU IL6 SIGNALING SCAR UP | 30 | 21 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |