Gene Page: VAX2
Summary ?
| GeneID | 25806 |
| Symbol | VAX2 |
| Synonyms | DRES93 |
| Description | ventral anterior homeobox 2 |
| Reference | MIM:604295|HGNC:HGNC:12661|Ensembl:ENSG00000116035|HPRD:05051|Vega:OTTHUMG00000129714 |
| Gene type | protein-coding |
| Map location | 2p13 |
| Pascal p-value | 0.963 |
| Fetal beta | -0.297 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20267441 | 2 | 71127363 | VAX2 | 9.8E-5 | -0.265 | 0.028 | DMG:Wockner_2014 |
| cg01719210 | 2 | 71128191 | VAX2 | 4.91E-4 | -0.263 | 0.047 | DMG:Wockner_2014 |
| cg15423212 | 2 | 71115225 | VAX2 | 3.54E-8 | -0.016 | 1.03E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
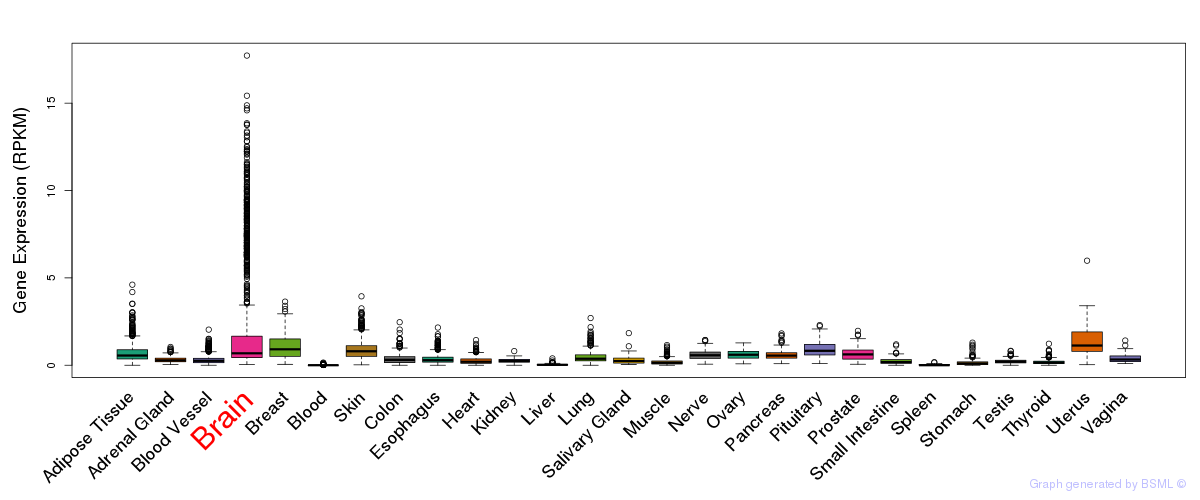
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UBE4B | 0.96 | 0.96 |
| DYRK1A | 0.96 | 0.96 |
| HERC1 | 0.95 | 0.95 |
| HUWE1 | 0.95 | 0.96 |
| CKAP5 | 0.95 | 0.96 |
| GON4L | 0.95 | 0.96 |
| KIAA0947 | 0.95 | 0.96 |
| KIAA1409 | 0.95 | 0.95 |
| RPRD2 | 0.95 | 0.96 |
| TP53BP1 | 0.94 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.81 | -0.85 |
| HIGD1B | -0.79 | -0.86 |
| FXYD1 | -0.79 | -0.83 |
| MT-CO2 | -0.79 | -0.84 |
| IFI27 | -0.78 | -0.84 |
| AF347015.21 | -0.76 | -0.87 |
| AF347015.33 | -0.76 | -0.78 |
| AF347015.27 | -0.75 | -0.79 |
| AF347015.8 | -0.75 | -0.81 |
| S100A16 | -0.74 | -0.79 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | NAS | 10485894 | |
| GO:0031490 | chromatin DNA binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007409 | axonogenesis | IEA | neuron, axon, neurite (GO term level: 12) | - |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009950 | dorsal/ventral axis specification | IEA | - | |
| GO:0007398 | ectoderm development | TAS | 10485894 | |
| GO:0048048 | embryonic eye morphogenesis | IEA | - | |
| GO:0007601 | visual perception | TAS | 10485894 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0043010 | camera-type eye development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |